Difference between revisions of "P1 Lab Activities"
| (50 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
[[File:Hackuarium_logo15.png|200px|thumb|]] | [[File:Hackuarium_logo15.png|200px|thumb|]] | ||
| − | + | Our P1 lab in Ecublens is the place where Hackuarium members can do synthetic biology and more, depending on risk analyses and plans. [https://drive.google.com/file/d/1tD2egvEsSemr7c4-DSdV0oi7O3rml2Ub/view?usp=sharing Here] are info for the current users, and your information... More documentation of our activities can be found linked from this current page, and the [[LaboP1 EN|most recent around the P1]] work.. | |
| − | + | Of course, biosafety is a key interest, and all users abide by the [https://diybio.org/codes/code-of-ethics-north-america-congress-2011/ ethical code]* of our open science community lab. | |
| − | |||
| − | + | ''Notre laboratoire P1 à Ecublens est l'endroit où les membres de Hackuarium peuvent faire de la biologie synthétique et plus, en fonction des analyses de risque et des plans. [https://drive.google.com/file/d/1tD2egvEsSemr7c4-DSdV0oi7O3rml2Ub/view?usp=sharing Voici des informations] pour les utilisateurs actuels, et vos informations... Plus de documentation sur nos activités peut être trouvée sur cette page, et la [[LaboP1|plus récente sur notre travail en laboratoire P1]].'' | |
| − | + | ''Bien sûr, la biosécurité est un intérêt majeur, et tous les utilisateurs respectent [https://diybio.org/codes/code-of-ethics-north-america-congress-2011/ le code éthique]* de notre laboratoire communautaire de science ouverte.'' | |
| − | '' | + | <br>* Here is an example of the code, DIYbio,(even if these days we prefer DIT (for Do-It-Together) research. / ''Voici un exemple du code de 'diy-bio' (même si en ces jours nous privilégions l'idée de faire ensemble) :'' |
| + | |||
| + | Respect | ||
| + | |||
| + | ''Respecter les êtres humains et tous les systèmes vivants.'' | ||
| − | |||
=Inspiration= | =Inspiration= | ||
| − | Several Hackuarium members | + | Several Hackuarium members had been working towards the goal of having a certified P1 laboratory since the foundation of the association in the spring of 2014. |
| + | |||
| + | In particular Yann P, Sam S and others did great leg-work, and even installed a 'green house' enclosure in the former open space (prior to renovation of UniverCité's second floor space). The work leading up to us finally getting the lab up and running is documented in [http://wiki.hackuarium.ch/w/Pushing_the_P1 this page]. <br> | ||
| − | + | One catalyst to get the renovated laboratory set up well was the participation of Dan Hernandez and Thomas Jordon to the "[https://bio.academany.org | How To Grow Almost Anything]" (HTGAA) course organised by MIT professor George Church. Once we moved to Ecublens, we adapted to fit the available space. | |
| − | + | Of course, it was not the 100m2 we started out with; and we are still under the threat of expulsion if the coop space is eventually destroyed as planned. | |
| − | + | In the meantime, post-pandemic (we hope), we continue to try to make these biohacking adventures sustainable. | |
| − | = | + | =Key Aims= |
| − | + | To learn, have fun, and avoid problems! | |
| − | Rachel Aronoff | + | Let us know if you want to do P1 experiments!<br> |
| + | =History and Biosafety Officers= | ||
| + | |||
| + | Here is the [[LaboP1 EN|most recent wiki page]] attempting to pull together the latest status of the P1 Lab project at Hackuarium (also available in [[LaboP1|French]]*). | ||
| + | |||
| + | A P1 laboratory, allowing members to select for bacterial clones carrying plasmids they wanted to use or make, was officially functional at the old Hackuarium site in Renens between 1 November and 1 September 2018. | ||
| + | |||
| + | Moving to Ecublens in the context of a cooperative in February 2019, we kept aiming to support members' projects and to safely pursue research, even during the time of the Covid-19 pandemic, when (not so surprisingly, after our friend Guy ran a GMO Detective workshop for our 5th anniversary festivities), we even contributed to a special 'Corona Detective' project, also supported by the CRI and JOGL, leading to an open access publication about this method of molecular diagnostics.<br><br>Work towards making a Hackuarium lab for biosafey level 1 experimentation began very early on, after the founding of Hackuarium. | ||
| + | |||
| + | This [[Pushing the P1|wiki page]] has some more history in it, and [[P1 SOP|this has detailed standard operating procedures]] put together initially. One aspect that was particularly important to Hackuarium was to have trained biosafety officers to oversee the work done in the P1 laboratory.<br> | ||
| + | |||
| + | Rachel Aronoff, who had started a P2 lab for viral work on sensory perception (in mice) at the EPFL, thus became the biosafety officer, while Olivier Emery initially agreed to take on the role of deputy biosafety officer. Later, Javier Ortiz took his place in this role.<br> | ||
If you need to contact them in particular about any proposed or in progress P1 activities, this is the best email address: biolab@hackuarium.ch <br> | If you need to contact them in particular about any proposed or in progress P1 activities, this is the best email address: biolab@hackuarium.ch <br> | ||
| − | + | Many people have already taken the "Introduction to biosafety" training necessary for full access to the P1 space. This has now been updated, and '''more training sessions can be organised upon request.''' | |
| − | <br> | + | Our P1 lab was inspected by representatives of the federal office for the environment in 2019 and in 2023, which probably have helped improve things greatly. |
| + | |||
| + | ''Notre laboratoire P1 était inspecté par les mandataires de l'office fédéral de l'environnement (OFEV) en 2019 et en 2023, qui ont probablement contribué à améliorer grandement les choses..''<br> | ||
| + | |||
| + | <nowiki>*</nowiki>This page (and others around biosafety and the P1 lab), probably still need more updates and many are still lacking most French translations. '''If you are interested in helping, please reach out!'''<br> | ||
=P1 experiments at Hackuarium= | =P1 experiments at Hackuarium= | ||
| − | Initially, these | + | Initially, these were all limited to bacterial growth of plasmid constructs**. <br> |
| − | Both classical and 'synthetic biology' strategies are possible! | + | Both classical and 'synthetic biology' strategies are possible! (Please see [[Imre's SynBio recources for advanced cloners|Imre's guide]] for more info in regards to the latter!) |
| + | |||
| + | We have already done experiments with various fluorescent plasmids and other expression plasmids (open insulin, N-gene for Covid-19 controls for our CoronaDetective), and did a special set of tests with antibiotic disks (Antibiogo) and simple ''E. coli'', but so far we have not yet cloned any new plasmids in the lab, as predicted back when Sam and Yann were planning ahead for the P1. However, notification to OFEV of a ''C. elegans'' crispr collaboration, to study a gene for quality control of gene expression with colleagues at Biozentrum (Basel), was additionally made in 2022, and 'transgenic animals' have thus also been experimented upon in the lab (but their parents were injected back up in Basel). | ||
| + | |||
| + | As we sometimes use a -80 freezer and other equipment also at the UNIL DMF, another SOP for transfer of OGMs has been made and validated. | ||
| + | |||
| + | |||
| − | + | <nowiki>**</nowiki> 'Construct' is the old school term for a plasmid, and 'to construct' is the proper verb for this process, btw, even if many people like to talk about cloning when putting bits of DNA together.<br> | |
==List of P1 Experiments== | ==List of P1 Experiments== | ||
| − | ''Make sure to link your project pages here!'' <br> | + | ''Make sure to link your project pages here!'' <br>All users in the P1 space are encouraged to use the on-line notebooks in Evernote, in order to help document their experiments. This method also allows ready sharing of protocols. Plasmid or other sequence information can also be readily analysed using the Benchling on-line application, and is recommended. |
| − | (more to come | + | (more to come ! ) <br> |
| − | [http://wiki.hackuarium.ch/w/Normalfreezer_expt DH5alpha test] | + | A [http://wiki.hackuarium.ch/w/Normalfreezer_expt DH5alpha test] began this adventure, with first test streaks done late in 2017... <br> |
| − | Bioluminescent strain growth <br> | + | Bioluminescent strain growth (turned out our presumptive ''Vibrio fischeri'' was actually ''Photobacterium phosphoreum''!)<br> |
| − | Mixed biosensor strain (containing 2 plasmids, an arsenic inducible GFP and constitutive mCherry)<br> | + | Mixed biosensor strain (containing 2 plasmids, an arsenic inducible GFP and constitutive mCherry from DMF-UNIL * see images below)<br> |
HTGAA set [http://wiki.hackuarium.ch/w/HTGAA in the old space especially!]<br> | HTGAA set [http://wiki.hackuarium.ch/w/HTGAA in the old space especially!]<br> | ||
[[OpenInsulin|Open Insulin]] | [[OpenInsulin|Open Insulin]] | ||
| − | [[Corona Detective]] | + | [[Corona Detective]] (To note, this molecular diagnostic method was an open science collaborative effort, and was mainly tested on control DNAs and RNAs, but for rare saliva use, these were collected and inactivated by the individual testing them, to reduce risks in the P1 lab. We gratefully acknowledge support from JOGL and its partners for this work.) |
| − | smg-4 revisited (''C. elegans'' gene for [[QualityControl4Genes|quality control of gene expression]]) | + | ''smg-4'' revisited (studies on a ''C. elegans'' gene for a type of [[QualityControl4Genes|quality control of gene expression]]). It includes attempts to rescue mutants with PCR products and to modify the gene in its normal location, in collaboration with the Mango lab at the Biozentrum in Basel. |
| + | Montreux Clean Beach project [[Montreux 2023|water analyses]], from EasyGel plates and ultimately cards, to classic microbial media (like Levine media), to the latest card format used in 2022/23... The colonies from these cards and plates were never cultivated further, even if used for molecular biological confirmation of the species identity (as published in our [https://besjournals.onlinelibrary.wiley.com/doi/10.1002/2688-8319.12094 open access article]). | ||
| − | + | & a project being developed currently (November 2023) | |
| − | + | - Cell Free Synthesis with plant extracts (So far, the plan is to avoid GMOs for this project...) | |
| − | |||
| − | + | Have something to add about '''your''' project idea?<br> | |
| − | + | or even other projects?? | |
| + | ==SOPs and Validation Experiments== | ||
| + | |||
| + | Here is the page begun by Sam and Yann in Renens: [http://wiki.hackuarium.ch/w/P1_SOP P1 standard operating procedure], which still has a great deal of good information, even if erroneous in some points. | ||
| − | + | Many experiments would benefit from more documentation and standard validated methods, as more keeps being learned. For instance, for sterilisation of media with the pressure cooker, we have learned that bacterial media is simpler to sterilise than substrates for mycelial growth, which needs more time! (Instead of just 20min at pressure for LB, for instance, mycelia substrate is better after at least 45mins.) | |
| + | |||
| + | Nonetheless, [https://drive.google.com/drive/folders/19hquLFedG5I9lh2Ni19sCy1Hw8qjrqFz?usp=sharing more SOPs are now put together], and laminated printouts of this information can furthermore be found in the 'Instructions' box on the shelf in the lab. | ||
| + | |||
| + | '''Standard Operation Procedure for inactivation of genetically modified organisms (GMOs) - bilingual FR/EN''' | ||
| + | |||
| + | Formal validation of the above SOP for GMO inactivation for disposal has now been done using commercial bleach on both fluorescent bacteria, and on nematodes transformed with the a plasmid, with good results. | ||
| − | + | To summarise briefly, each set of experiments tested how many surviving GMOs would be found after inactivation tests with 100% and 10% bleach, or just water. In both cases, bacteria or worms, the 10% solutions were sufficient to prevent further growth of the GMOs. However, using the 100% solution is a bit more safe, as some contaminants, like spores from molds, might only be inactivated at higher concentrations. | |
| − | + | For details: please see [[Report ValidTestsGMOinact|the report]] and more [[Validated GMO inactivation|visual summary]], both also available in French versions ([[Rapport inactOGM|rapport]] & résumés sur les validations avec [[OGMinact validation|bactéries]] et [[Validations nematodes|nématodes]]) and you can also [https://www.evernote.com/shard/s586/sh/9e3be51e-434c-6b7d-25ed-ecb34b5bfe2a/HNkV4W3AW3UMJNqd7--KWVclJ5uwYYFC3Xq7oAL2MPJkgITpTeUvG5wErw see Evernote with details for the bacterial] and [https://www.evernote.com/shard/s586/sh/4ff409d3-1754-4d1f-8507-ea7780f1b1c4/cwK7hplOCDHfUdnjJhScKxOfGgGzVJDRz9nDTJY1vrBHYZkT2atVzeDnFQ for the transformed worms] tests. | |
| + | * | ||
| − | Here is the [https:// | + | <br> |
| + | =Timeline for Project Initiation= | ||
| + | 1) Talk with everyone about your ideas and what you would like to do.<br> | ||
| + | 2) Take the safety course and do risk assessment on your idea. Here is the [https://prezi.com/view/f0XXqskwRPwMuiP9vrx0/ updated prezi used for this now, 2023], and here is the form to help document your risk assessment results.<br> | ||
| + | 3) Start documenting your idea in the wiki. You can use [http://wiki.hackuarium.ch/w/Hypo_proj this template] developed by YannP as a guide. <br> | ||
| + | 4) Fill in [http://wiki.hackuarium.ch/w/Form:GMO_Project the form], also developed by YannP, especially if you plan to do any plasmid cloning or transformation! <br> | ||
| + | 5) Get going! (ask for a minigrant, if necessary for obtaining reagents or other materials.) <br> | ||
| − | + | <br> | |
<br> | <br> | ||
| Line 91: | Line 132: | ||
The first images are of plates carrying our first plasmid strains, streaked at UNIL and grown in our own incubator, and of Vlad streaking the bioluminescent strain... <br> The big -80 freezer behind him can hold our stock, at DMF UNIL, until we know if the -20 method will work, or are more fully equipped! <br> | The first images are of plates carrying our first plasmid strains, streaked at UNIL and grown in our own incubator, and of Vlad streaking the bioluminescent strain... <br> The big -80 freezer behind him can hold our stock, at DMF UNIL, until we know if the -20 method will work, or are more fully equipped! <br> | ||
| − | To note: The bioluminescent strain is supposed to be moderately expressed, but on the plate after overnight incubation at 37<sup>o</sup>C, almost no light is visible by eye... <br> | + | To note: The bioluminescent strain is supposed to be moderately expressed, but on the plate after overnight incubation at 37<sup>o</sup>C, almost no light is visible by eye... <br> |
| − | A liquid culture | + | A liquid culture was also attempted next, with the same effect...<br> |
| − | Vlad helpfully found it in the DMF collection, but more work is planned to make it bright! <br> | + | Vlad helpfully found it in the DMF collection, but more work is planned to make it bright! (Ultimately, we learned that time at in the refrigerator at 4 degrees celsius was necessary for the ''P. phosphoreum'' to produce visible light! The aldehyde substrate necessary for the light reaction, but also potentially toxic to the bugs carrying it, means that the full operon is under strict quorum regulation...)<br> |
The second strain actually carries two plasmids, a constitutive mCherry one, and an arsenic inducible GFP one... The experiment being tested is whether, by plating this strain on the two different antibiotics, new sub-strains can be selected carrying each plasmid alone... (we already have stocks of the parent strain in the UNIL freezer! It was actually made by Rachel for her 'nematode assisted biosensor analyses' from a few years back!)<br> | The second strain actually carries two plasmids, a constitutive mCherry one, and an arsenic inducible GFP one... The experiment being tested is whether, by plating this strain on the two different antibiotics, new sub-strains can be selected carrying each plasmid alone... (we already have stocks of the parent strain in the UNIL freezer! It was actually made by Rachel for her 'nematode assisted biosensor analyses' from a few years back!)<br> | ||
| − | There are a couple more images for the test of bacterial strain stability in the -20 freezer, with the DH5alpha strain frozen in either glycerol vs milk. This isn't strictly a P1 experiment, because these bacteria were not selected and carry no plasmids... If they are stable, however, in other words, if they can be repeatedly propagated from this stock, this will mean our P1 lab may not need (even though it would be good if we were able to get one!) a -80 freezer, as is the usual standard for strain stocks... <br> | + | There are a couple more images for the test of bacterial strain stability in the -20 freezer, with the DH5alpha strain frozen in either glycerol vs milk. This isn't strictly a P1 experiment, because these bacteria were not selected and carry no plasmids... If they are stable, however, in other words, if they can be repeatedly propagated from this stock, this will mean our P1 lab may not need (even though it would be good if we were able to get one!) a -80 freezer, as is the usual standard for strain stocks... <br>(To Note: We have come a long way since 2017, and we now know that many bacteria do survive in the -20 freezer from glycerol stocks, but the milk method gave more variable results. However, the nematodes were not at all happy, after being frozen in the -20... This leads directly to the next section of this page.) |
| − | + | = Wishlist for the P1 Lab = | |
| − | + | We always have known our coop space in Ecublens is destined to be destroyed, so we have made do in spite of some problematic issues (carpeting in the hallways, lack of a sink in the P1 itself, an overall need for more space, etc.). However, that has never prevented us from hoping for more good things to make everything better. | |
| + | |||
| + | Below, find some of the key items of our current wishlist. | ||
| + | |||
| + | * A -80 freezer to stock strains. | ||
| + | * A refrigerator that won't freeze up so often. | ||
| + | * A more reliable -20 freezer... | ||
| + | * A source for clean water for molecular microbiology.** | ||
| + | * A sink in the lab space. | ||
| + | * A laminar air flow hood, in the P1 lab. | ||
| + | * A space with more room for biomaterials, chemistry, workshops and more. | ||
| + | * | ||
| + | * | ||
| − | < | + | = Further information = |
| − | < | + | [[File:Newsafetyshelf.jpg|thumb|the new 'safety' shelf - with first aide, eyewash, fire-extinguisher, absorbant material, etc.]] |
| + | <big>Always be aware when working in the lab, and do not be in a rush!</big> | ||
| − | + | [[File:Safetyshelf.jpg|thumb|the old 'safety' shelf - with first aid, eye wash, googles, etc...]] | |
| − | + | [[File:Sterile technique.jpg|thumb|using the gas canister for liquid culture inoculations]] | |
| − | + | [[File:Top hood.jpg|thumb|extra info on the hood - now also printed nicely...]] | |
| − | + | <big>Additionally, our basic microbiology hood is not for GMOs !</big> | |
| − | |||
| − | |||
| − | + | (Classic aseptic technique around a flame in the lab is always a standard part of our intro to biosafety training, but let us know if you have any questions, before initiating your experiment. We have made a collection of SOPs, now, including how to use the gas canister for sterile technique inside the P1 itself.) | |
| − | = | + | * Here is our initial [http://wiki.hackuarium.ch/w/P1_SOP P1 standard operating procedure]. More historical perspective is pulled together now also in |
| + | * General Community Lab biosafety information is also [https://communitybio.pubpub.org/pub/m255th2q/release/1 available on line]. | ||
| + | * From our pandemic experiences, [https://docs.google.com/document/d/1ldQ1aRxHjuj-U7kNqwWlkwwwW54U6ls8xMI9l0LBFrQ/edit?usp=sharing here is another good starting point]. | ||
| + | *Here is the [https://drive.google.com/open?id=0B1jaPHOkH_42dVhjOXBBN0xsUms link to all the initial P1 safety info, including our list of users and list of strains] for future reference. | ||
| + | *Here is the [[LaboP1 EN|latest page]] pulling together most all the P1 lab info from this wiki.<br> | ||
<br> | <br> | ||
| + | See something that needs mentioning? You can help contribute! | ||
| − | + | Come join us!<br> | |
| − | + | * Do you have questions? Maybe you can't reach Rachel or Javier, or want to find out more before even trying to ask them?? (While there is still an active link to an old service in the DIYBio community for this - [http://ask.diybio.org/ Ask a Biosafety Professional] - this may no longer be a functioning entity - our 2nov23 test has not received any response...) | |
| − | * Do you have questions? Maybe you can't reach Rachel or | + | * The [https://communitybio.pubpub.org/pub/m255th2q/release/1 community lab biosafety handbook] has very extensive details, thanks to colleagues around the world.<br> |
<br> | <br> | ||
'''Remember, being clueless is no excuse!!''' | '''Remember, being clueless is no excuse!!''' | ||
| − | |||
| − | |||
[[Category:Work In Progress]] | [[Category:Work In Progress]] | ||
Latest revision as of 10:31, 21 December 2023
Our P1 lab in Ecublens is the place where Hackuarium members can do synthetic biology and more, depending on risk analyses and plans. Here are info for the current users, and your information... More documentation of our activities can be found linked from this current page, and the most recent around the P1 work..
Of course, biosafety is a key interest, and all users abide by the ethical code* of our open science community lab.
Notre laboratoire P1 à Ecublens est l'endroit où les membres de Hackuarium peuvent faire de la biologie synthétique et plus, en fonction des analyses de risque et des plans. Voici des informations pour les utilisateurs actuels, et vos informations... Plus de documentation sur nos activités peut être trouvée sur cette page, et la plus récente sur notre travail en laboratoire P1.
Bien sûr, la biosécurité est un intérêt majeur, et tous les utilisateurs respectent le code éthique* de notre laboratoire communautaire de science ouverte.
* Here is an example of the code, DIYbio,(even if these days we prefer DIT (for Do-It-Together) research. / Voici un exemple du code de 'diy-bio' (même si en ces jours nous privilégions l'idée de faire ensemble) :
Respect
Respecter les êtres humains et tous les systèmes vivants.
Inspiration
Several Hackuarium members had been working towards the goal of having a certified P1 laboratory since the foundation of the association in the spring of 2014.
In particular Yann P, Sam S and others did great leg-work, and even installed a 'green house' enclosure in the former open space (prior to renovation of UniverCité's second floor space). The work leading up to us finally getting the lab up and running is documented in this page.
One catalyst to get the renovated laboratory set up well was the participation of Dan Hernandez and Thomas Jordon to the "| How To Grow Almost Anything" (HTGAA) course organised by MIT professor George Church. Once we moved to Ecublens, we adapted to fit the available space.
Of course, it was not the 100m2 we started out with; and we are still under the threat of expulsion if the coop space is eventually destroyed as planned.
In the meantime, post-pandemic (we hope), we continue to try to make these biohacking adventures sustainable.
Key Aims
To learn, have fun, and avoid problems!
Let us know if you want to do P1 experiments!
History and Biosafety Officers
Here is the most recent wiki page attempting to pull together the latest status of the P1 Lab project at Hackuarium (also available in French*).
A P1 laboratory, allowing members to select for bacterial clones carrying plasmids they wanted to use or make, was officially functional at the old Hackuarium site in Renens between 1 November and 1 September 2018.
Moving to Ecublens in the context of a cooperative in February 2019, we kept aiming to support members' projects and to safely pursue research, even during the time of the Covid-19 pandemic, when (not so surprisingly, after our friend Guy ran a GMO Detective workshop for our 5th anniversary festivities), we even contributed to a special 'Corona Detective' project, also supported by the CRI and JOGL, leading to an open access publication about this method of molecular diagnostics.
Work towards making a Hackuarium lab for biosafey level 1 experimentation began very early on, after the founding of Hackuarium.
This wiki page has some more history in it, and this has detailed standard operating procedures put together initially. One aspect that was particularly important to Hackuarium was to have trained biosafety officers to oversee the work done in the P1 laboratory.
Rachel Aronoff, who had started a P2 lab for viral work on sensory perception (in mice) at the EPFL, thus became the biosafety officer, while Olivier Emery initially agreed to take on the role of deputy biosafety officer. Later, Javier Ortiz took his place in this role.
If you need to contact them in particular about any proposed or in progress P1 activities, this is the best email address: biolab@hackuarium.ch
Many people have already taken the "Introduction to biosafety" training necessary for full access to the P1 space. This has now been updated, and more training sessions can be organised upon request.
Our P1 lab was inspected by representatives of the federal office for the environment in 2019 and in 2023, which probably have helped improve things greatly.
Notre laboratoire P1 était inspecté par les mandataires de l'office fédéral de l'environnement (OFEV) en 2019 et en 2023, qui ont probablement contribué à améliorer grandement les choses..
*This page (and others around biosafety and the P1 lab), probably still need more updates and many are still lacking most French translations. If you are interested in helping, please reach out!
P1 experiments at Hackuarium
Initially, these were all limited to bacterial growth of plasmid constructs**.
Both classical and 'synthetic biology' strategies are possible! (Please see Imre's guide for more info in regards to the latter!)
We have already done experiments with various fluorescent plasmids and other expression plasmids (open insulin, N-gene for Covid-19 controls for our CoronaDetective), and did a special set of tests with antibiotic disks (Antibiogo) and simple E. coli, but so far we have not yet cloned any new plasmids in the lab, as predicted back when Sam and Yann were planning ahead for the P1. However, notification to OFEV of a C. elegans crispr collaboration, to study a gene for quality control of gene expression with colleagues at Biozentrum (Basel), was additionally made in 2022, and 'transgenic animals' have thus also been experimented upon in the lab (but their parents were injected back up in Basel).
As we sometimes use a -80 freezer and other equipment also at the UNIL DMF, another SOP for transfer of OGMs has been made and validated.
** 'Construct' is the old school term for a plasmid, and 'to construct' is the proper verb for this process, btw, even if many people like to talk about cloning when putting bits of DNA together.
List of P1 Experiments
Make sure to link your project pages here!
All users in the P1 space are encouraged to use the on-line notebooks in Evernote, in order to help document their experiments. This method also allows ready sharing of protocols. Plasmid or other sequence information can also be readily analysed using the Benchling on-line application, and is recommended.
(more to come ! )
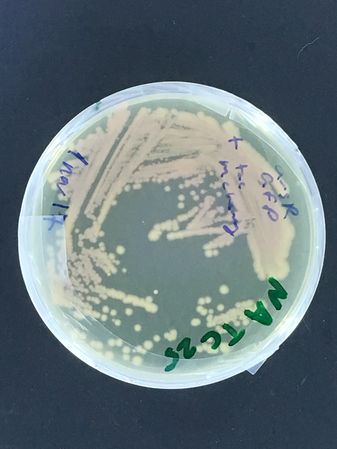
A DH5alpha test began this adventure, with first test streaks done late in 2017...
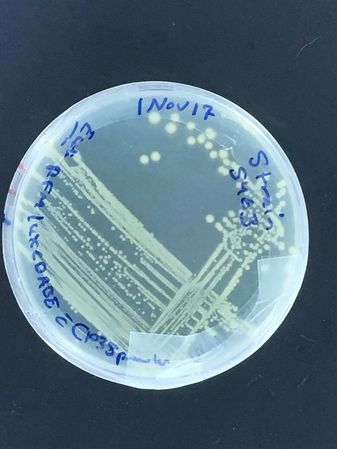
Bioluminescent strain growth (turned out our presumptive Vibrio fischeri was actually Photobacterium phosphoreum!)
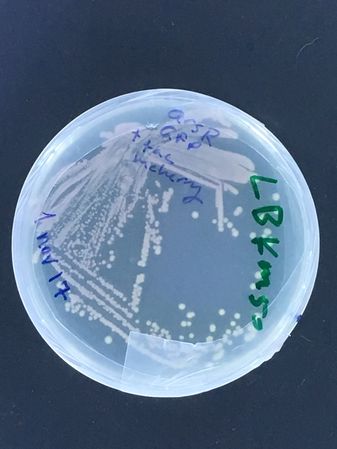
Mixed biosensor strain (containing 2 plasmids, an arsenic inducible GFP and constitutive mCherry from DMF-UNIL * see images below)
HTGAA set in the old space especially!
Open Insulin
Corona Detective (To note, this molecular diagnostic method was an open science collaborative effort, and was mainly tested on control DNAs and RNAs, but for rare saliva use, these were collected and inactivated by the individual testing them, to reduce risks in the P1 lab. We gratefully acknowledge support from JOGL and its partners for this work.)
smg-4 revisited (studies on a C. elegans gene for a type of quality control of gene expression). It includes attempts to rescue mutants with PCR products and to modify the gene in its normal location, in collaboration with the Mango lab at the Biozentrum in Basel.
Montreux Clean Beach project water analyses, from EasyGel plates and ultimately cards, to classic microbial media (like Levine media), to the latest card format used in 2022/23... The colonies from these cards and plates were never cultivated further, even if used for molecular biological confirmation of the species identity (as published in our open access article).
& a project being developed currently (November 2023)
- Cell Free Synthesis with plant extracts (So far, the plan is to avoid GMOs for this project...)
Have something to add about your project idea?
or even other projects??
SOPs and Validation Experiments
Here is the page begun by Sam and Yann in Renens: P1 standard operating procedure, which still has a great deal of good information, even if erroneous in some points.
Many experiments would benefit from more documentation and standard validated methods, as more keeps being learned. For instance, for sterilisation of media with the pressure cooker, we have learned that bacterial media is simpler to sterilise than substrates for mycelial growth, which needs more time! (Instead of just 20min at pressure for LB, for instance, mycelia substrate is better after at least 45mins.)
Nonetheless, more SOPs are now put together, and laminated printouts of this information can furthermore be found in the 'Instructions' box on the shelf in the lab.
Standard Operation Procedure for inactivation of genetically modified organisms (GMOs) - bilingual FR/EN
Formal validation of the above SOP for GMO inactivation for disposal has now been done using commercial bleach on both fluorescent bacteria, and on nematodes transformed with the a plasmid, with good results.
To summarise briefly, each set of experiments tested how many surviving GMOs would be found after inactivation tests with 100% and 10% bleach, or just water. In both cases, bacteria or worms, the 10% solutions were sufficient to prevent further growth of the GMOs. However, using the 100% solution is a bit more safe, as some contaminants, like spores from molds, might only be inactivated at higher concentrations.
For details: please see the report and more visual summary, both also available in French versions (rapport & résumés sur les validations avec bactéries et nématodes) and you can also see Evernote with details for the bacterial and for the transformed worms tests.
Timeline for Project Initiation
1) Talk with everyone about your ideas and what you would like to do.
2) Take the safety course and do risk assessment on your idea. Here is the updated prezi used for this now, 2023, and here is the form to help document your risk assessment results.
3) Start documenting your idea in the wiki. You can use this template developed by YannP as a guide.
4) Fill in the form, also developed by YannP, especially if you plan to do any plasmid cloning or transformation!
5) Get going! (ask for a minigrant, if necessary for obtaining reagents or other materials.)
Gallery
The first images are of plates carrying our first plasmid strains, streaked at UNIL and grown in our own incubator, and of Vlad streaking the bioluminescent strain...
The big -80 freezer behind him can hold our stock, at DMF UNIL, until we know if the -20 method will work, or are more fully equipped!
To note: The bioluminescent strain is supposed to be moderately expressed, but on the plate after overnight incubation at 37oC, almost no light is visible by eye...
A liquid culture was also attempted next, with the same effect...
Vlad helpfully found it in the DMF collection, but more work is planned to make it bright! (Ultimately, we learned that time at in the refrigerator at 4 degrees celsius was necessary for the P. phosphoreum to produce visible light! The aldehyde substrate necessary for the light reaction, but also potentially toxic to the bugs carrying it, means that the full operon is under strict quorum regulation...)
The second strain actually carries two plasmids, a constitutive mCherry one, and an arsenic inducible GFP one... The experiment being tested is whether, by plating this strain on the two different antibiotics, new sub-strains can be selected carrying each plasmid alone... (we already have stocks of the parent strain in the UNIL freezer! It was actually made by Rachel for her 'nematode assisted biosensor analyses' from a few years back!)
There are a couple more images for the test of bacterial strain stability in the -20 freezer, with the DH5alpha strain frozen in either glycerol vs milk. This isn't strictly a P1 experiment, because these bacteria were not selected and carry no plasmids... If they are stable, however, in other words, if they can be repeatedly propagated from this stock, this will mean our P1 lab may not need (even though it would be good if we were able to get one!) a -80 freezer, as is the usual standard for strain stocks...
(To Note: We have come a long way since 2017, and we now know that many bacteria do survive in the -20 freezer from glycerol stocks, but the milk method gave more variable results. However, the nematodes were not at all happy, after being frozen in the -20... This leads directly to the next section of this page.)
Wishlist for the P1 Lab
We always have known our coop space in Ecublens is destined to be destroyed, so we have made do in spite of some problematic issues (carpeting in the hallways, lack of a sink in the P1 itself, an overall need for more space, etc.). However, that has never prevented us from hoping for more good things to make everything better.
Below, find some of the key items of our current wishlist.
- A -80 freezer to stock strains.
- A refrigerator that won't freeze up so often.
- A more reliable -20 freezer...
- A source for clean water for molecular microbiology.**
- A sink in the lab space.
- A laminar air flow hood, in the P1 lab.
- A space with more room for biomaterials, chemistry, workshops and more.
Further information
Always be aware when working in the lab, and do not be in a rush!
Additionally, our basic microbiology hood is not for GMOs !
(Classic aseptic technique around a flame in the lab is always a standard part of our intro to biosafety training, but let us know if you have any questions, before initiating your experiment. We have made a collection of SOPs, now, including how to use the gas canister for sterile technique inside the P1 itself.)
- Here is our initial P1 standard operating procedure. More historical perspective is pulled together now also in
- General Community Lab biosafety information is also available on line.
- From our pandemic experiences, here is another good starting point.
- Here is the link to all the initial P1 safety info, including our list of users and list of strains for future reference.
- Here is the latest page pulling together most all the P1 lab info from this wiki.
See something that needs mentioning? You can help contribute!
Come join us!
- Do you have questions? Maybe you can't reach Rachel or Javier, or want to find out more before even trying to ask them?? (While there is still an active link to an old service in the DIYBio community for this - Ask a Biosafety Professional - this may no longer be a functioning entity - our 2nov23 test has not received any response...)
- The community lab biosafety handbook has very extensive details, thanks to colleagues around the world.
Remember, being clueless is no excuse!!