Difference between revisions of "AGiR! for genomic integrity"
| (95 intermediate revisions by 4 users not shown) | |||
| Line 1: | Line 1: | ||
[[File:MinilogoB.jpg |200px|thumb|]] | [[File:MinilogoB.jpg |200px|thumb|]] | ||
| − | Genomic integrity is a | + | Genomic integrity is a 'big picture' concept to aid public health efforts and basically includes all the molecular genetic details of cells. |
| − | So many things we commonly choose to do can impact genomic integrity, also potentially | + | So many things we commonly choose to do can impact genomic integrity, also potentially affecting future generations. <br> |
| − | At | + | '''To Note: the concept of 'dynamic genomic integrity' celebrated its 15th anniversary already.''' <br> |
| − | <br> | + | It was first proposed in 2009, as part of the old EFV Prize (to help End Fashion Victimisation)... |
| − | Here is the basic plan: using inner cheek cells for micronuclei and comet assays! | + | |
| − | <br> | + | (AGiR! as a Swiss Association began over 11 years ago, with the first general assembly meeting held on 16sept2013)<br> |
| − | Watch this space for more info and visit | + | |
| − | [http://www.genomicintegrity.org | + | At Hackuarium, AGiR! hopes to get participative research actions really going - developing DIT research methods to readily assay *your very own* cells for DNA damage, using classic (circa 1980s) techniques. If these can already be sold in 'kits,' maybe it will also be simple to make them available to all for open source efforts?<br> |
| + | |||
| + | Here is the basic plan: using inner cheek cells for micronuclei and comet assays, because there are still many things we can choose in life, to help protect genomic integrity, even if this is only one way to measure it (actual DNA breaks)! <br> Inspired by GOSH2018, here also is the AGiR! summary flyer in Chinese [[File:AGIRFlyerChinese_smaller.jpg|200px|thumb|left| AGiR! summary flyer in Chinese]](left), to help everyone think about these issues.<br> | ||
| + | |||
| + | If you want the quick idea, without reading too much more, the [https://drive.google.com/open?id=0B1jaPHOkH_42YmlhUThkNy1aN0U poster explaining genomic integrity projects at Hackuarium] is worth checking out!<br> | ||
| + | |||
| + | Watch this space for more info and visit the official website of [http://www.genomicintegrity.org AGiR! Association]<br> | ||
| + | AGiR is also carrying out the following projects in the Hackuarium community laboratory:<br> | ||
| + | * [[Moss Menageries with AGiR!]] | ||
| + | * [[Magnesium tests with AGiR!]] | ||
| + | * [[Bioluminescence investigations]] | ||
| + | |||
| + | and, [http://interspecifics.cc/comunicacionesespeculativas/media/TL10%20.mp4?fbclid=IwAR21X5tyOei3Aupwb5O3reVfHjnX9RP0sKufG6IMLJzqsC4XvKAOWE4IOTA if this inspires you], what do you think about joining in on a new [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2268691/ project about swarming bacteria] like these?? (potentially!)<br> | ||
| + | <br> | ||
== Introduction == | == Introduction == | ||
=== Context === | === Context === | ||
| − | This is a part of a | + | This is a part of a Hackuarium project by AGiR! on [[genomic integrity]], to find out more and help raise awareness in the public interest. <br> |
This wiki page is under construction, and will help keep track as we develop open source simple protocols of two classic methods for quantitation of DNA damage in human cells, the micronuclei and comet assays. | This wiki page is under construction, and will help keep track as we develop open source simple protocols of two classic methods for quantitation of DNA damage in human cells, the micronuclei and comet assays. | ||
=== People === | === People === | ||
'''Contact and lead:''' | '''Contact and lead:''' | ||
| − | *Rachel Aronoff - Founder and | + | *Rachel Aronoff - Founder and CSO of AGiR! Action for Genomic integrity through Research! |
'''Participants:''' | '''Participants:''' | ||
| − | *maybe you?! | + | * AGiR! members |
| + | * Damian Jurkiewicz | ||
| + | * Tatiana Eliseeva | ||
| + | * Anna Dussuet | ||
| + | * Violaine Regard | ||
| + | * Jennifer Veillard | ||
| + | * Vithooban Thavapalan | ||
| + | * maybe you?! | ||
=== Main Goals === | === Main Goals === | ||
| − | Genomic integrity is a big picture concept which includes basically all the molecular genetic details of cells (from RNAs to DNAs and the epigenetic marks for proper gene expression), and thus is a topic that can be explored in several ways. For the genomic integrity project at the Hackuarium the focus will be on directly quantifying DNA damage. Comet and micronuclei assays were developed in the 1980s and publications about use of these assays on human cheek cells are already in the literature. These methods, as open source protocols, will encourage citizen science and raise awareness. Additionally, they can provide an easily "workshopable" way to show people more about their own cells, while learning how to assess DNA damage.<br> In other words, this experiment can be used as a gateway, to demonstrate and raise awareness for everyone on the damage possible from many common substances or activities - on '''your own cells, your own genetic information'''!<br> In the scope of Hackuarium activities, mammalian cell cultures are still (april 2016) a no go, but freshly isolated cheek cells from individuals can be used to do tests on DNA damaging compounds or activities. Such 'ex situ' assays might seem more attractive, especially if potentially toxic compounds are tested! (but the caveat is that it is only after the metabolism of some compounds that the DNA-damaging effects are 'revealed' - for example alcohol to DNA-damaging aldehydes…) | + | Genomic integrity is a big picture concept which includes basically all the molecular genetic details of cells (from RNAs to DNAs and the epigenetic marks for proper gene expression), and thus is a topic that can be explored in several ways. For the genomic integrity project at the Hackuarium the focus will be on directly quantifying DNA damage. Comet and micronuclei assays were developed in the 1980s and publications about use of these assays on human cheek cells are already in the literature. These methods, as open source protocols, will encourage citizen science and raise awareness. Additionally, they can provide an easily "workshopable" way to show people more about their own cells, while learning how to assess DNA damage.<br> In other words, this experiment can be used as a gateway, to demonstrate and raise awareness for everyone on the damage possible from many common substances or activities - on '''your own cells, your own genetic information'''!<br> In the scope of Hackuarium activities, mammalian cell cultures are still (april 2016) a no go, but freshly isolated cheek cells from individuals can be used to do tests on DNA damaging compounds or activities. Such 'ex situ' assays might seem more attractive, especially if potentially toxic compounds are tested! (but the caveat is that it is only after the metabolism of some compounds that the DNA-damaging effects are 'revealed' - for example alcohol to DNA-damaging aldehydes…). For this reason, 'citizen scientists' will be encouraged to first get baseline measures on their cells, then do a 'trial' and test its effects (i.e. daily swimming, chocolate or blueberry consumption, or some other activity). The comet and micronuclei assay results can also be compared after a 'wash-out' stage, without the given trial activity… These would all provide 'biomonitoring' results about the given 'intervention' from baseline conditions. <br> Another possibility with these assays are to perform ''ex situ'' experiments on your own cells, as mentioned, which would be giving acute genotoxicity information... <br> In order to further investigate compounds that might cause DNA damage, a classic test of bacterial reversion (i.e. the Ames test) or on bulk DNA may also be nice to establish at the Hackuarium. <br> If enough people follow the 'open source' protocol(s) that we develop to follow controlled exposure protocols or test substances directly on their isolated cheek cells, and clearly significant effects are observed, we may even be able to corroborate or refute previous works in the literature. (Please go to the bottom of the page, for one concrete example being considered.) <br> Your ideas and comments are welcome throughout btw! |
=== Objectives === | === Objectives === | ||
| Line 32: | Line 52: | ||
* Obtain ready results with cheek cells for the comet and micronuclei assays | * Obtain ready results with cheek cells for the comet and micronuclei assays | ||
| − | * Put together the open source | + | * Put together the open source protocols for each method, with suggestions for standard tests and controls |
* Demonstrate methods and 'share the love' with other biohacker groups (La Paillasse etc.) | * Demonstrate methods and 'share the love' with other biohacker groups (La Paillasse etc.) | ||
| − | * Study the feasibility of further methods that could contribute to this project - i.e. | + | * Study the feasibility of further methods that could contribute to this project - i.e. open-source imaging and microscopy techniques, like the [https://openflexure.org/ OpenFlexure] system, which we are trying for imaging comets by epifluorescence. As of 2019 we had gotten to the point of seeing fluor nuclei, but not comet tails, but then the pandemic caused us to put this to the side, to focus on Corona Detective R&D. In 2022, we hoped to get back to all this 'cheek cell chip' work, and it is still of great interest! '''You''' could help! |
| − | |||
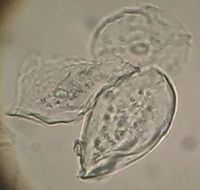
| − | '''Cheek cells are continually renewing epithelial cells and are about 50 micrometers in diameter.''' [[File:Cells3.jpg|200px|thumb|right|Three cheek cells imaged on the scope at the Hackuarium | + | '''Cheek cells are continually renewing epithelial cells and are about 50 micrometers in diameter.''' [[File:Cells3.jpg|200px|thumb|right|Three cheek cells imaged on the scope at the Hackuarium. ]] |
== Background & Inspiration == | == Background & Inspiration == | ||
| Line 47: | Line 66: | ||
=== Some theory === | === Some theory === | ||
More on comet cells and micronuclei to come soon! | More on comet cells and micronuclei to come soon! | ||
| − | + | However, in the meantime, here are the references to the cheek cell papers mentioned in the presentation (and in the prezi): <br> | |
Szeto et al (2005) Mutation Research 578:371-381<br> | Szeto et al (2005) Mutation Research 578:371-381<br> | ||
(In this paper, green tea is also shown to be protective.)<br> | (In this paper, green tea is also shown to be protective.)<br> | ||
dos Santos Rocha et al (2014) Genetics and Molecular Biology, 37(4)702-707<br> | dos Santos Rocha et al (2014) Genetics and Molecular Biology, 37(4)702-707<br> | ||
(This paper looks at micronuclei in cheek cells (and also analyses apoptosis) from mouthwash and alcohol users.)<br> | (This paper looks at micronuclei in cheek cells (and also analyses apoptosis) from mouthwash and alcohol users.)<br> | ||
| + | Also here is a [https://www.sciencedirect.com/science/article/pii/S1383574218300620?via%3Dihub new review about micronuclei]! | ||
''To note: the two assays proposed would be analysing only limited aspects of 'genomic integrity' at the DNA level - large scale chromosomal disruption in the micronuclei assay and double-stranded breaks with the comet assay! <br>There are many more methods that could look at other aspects (particular nucleotide lesions, reactive oxygen species and inflammation, gene expression, microRNAs, etc.) of genomic integrity!'' | ''To note: the two assays proposed would be analysing only limited aspects of 'genomic integrity' at the DNA level - large scale chromosomal disruption in the micronuclei assay and double-stranded breaks with the comet assay! <br>There are many more methods that could look at other aspects (particular nucleotide lesions, reactive oxygen species and inflammation, gene expression, microRNAs, etc.) of genomic integrity!'' | ||
| Line 59: | Line 79: | ||
* [https://exogenbio.com/] Sylvain Costes and Jon Tang made this great effort in 2014, based upon sub-cellular fluorescence quantification of DNA Damage Response components, but their blood draw kit was a bit tricky for some, and Sylvain told me the results were not so reliable after these fixed cells arrived in the mail, as opposed to their results when they had them 'fresh.' | * [https://exogenbio.com/] Sylvain Costes and Jon Tang made this great effort in 2014, based upon sub-cellular fluorescence quantification of DNA Damage Response components, but their blood draw kit was a bit tricky for some, and Sylvain told me the results were not so reliable after these fixed cells arrived in the mail, as opposed to their results when they had them 'fresh.' | ||
* [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4246922/] This is a Frontiers in Genetics article with a nice focus on using inner cheek cells and other epithelial cells for DNA damage assessment, also from 2014. | * [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4246922/] This is a Frontiers in Genetics article with a nice focus on using inner cheek cells and other epithelial cells for DNA damage assessment, also from 2014. | ||
| + | * [http://www.sciencedirect.com/science/article/pii/S138357181300003X This paper] describes results of DNA damage scoring in soy bean farm workers exposed to various pesticides, with DNA damage scored via comet assays of blood cells and micronuclei assays of cheek cells. | ||
| + | |||
| + | == Protocol (Plan) == | ||
| + | To do both comet cell and micronuclei assays by the simplest possible methods, with the bare minimal laboratory reagents/infrastructure... | ||
| + | To potentially get a 'cheek cell chip' working - so that micronuclei scoring then the comet assay will allow one to assess the level of damage for all the cells in the sample for both methods... | ||
=== Setup description === | === Setup description === | ||
General principle for both assays: looking at treated cells on slides through a microscope, imaging and quantitating the effects (i.e. counting number of cells with micronuclei relative to the total number of cells, or determining the length of the 'comet tail' - the longer the 'tail' the more DNA breaks - on average). <br> | General principle for both assays: looking at treated cells on slides through a microscope, imaging and quantitating the effects (i.e. counting number of cells with micronuclei relative to the total number of cells, or determining the length of the 'comet tail' - the longer the 'tail' the more DNA breaks - on average). <br> | ||
| − | The comet assay requires more treatments and a short 'electrophoresis' step, as compared to the micronuclei assay. | + | The comet assay requires more treatments and a short 'electrophoresis' step, as compared to the micronuclei assay. <br> |
| + | For the purpose of cheek cell comet assays, use of a special chamber with mini-electrophoresis capacity, capable of microscopic investigation, is being planned. | ||
=== Necessary Materials === | === Necessary Materials === | ||
| Line 73: | Line 99: | ||
:::* Mercury Lamp Setup and filters for fluorescent dye detection. | :::* Mercury Lamp Setup and filters for fluorescent dye detection. | ||
:::* Could also try to design a transilluminator suitable for cellular resolution (UV/Fluo as usual for comets) when imaging (to look for open source models) | :::* Could also try to design a transilluminator suitable for cellular resolution (UV/Fluo as usual for comets) when imaging (to look for open source models) | ||
| − | + | * Other reagents and histochemical stains (methylene blue) | |
| − | + | * Hydrogen peroxide for positive control cells (oxidative damage) | |
| − | + | * Antioxidant molecules as possible 'protective' intervention . (i.e. green tea as in Szeto et al...) | |
| − | + | <nowiki>*</nowiki>Proteinase K is recommended in buccal cell protocols, but DIY alternatives are being considered, like tablets for contact lens cleaning <br> | |
| − | |||
| − | |||
'** To protect the DNA from digestion | '** To protect the DNA from digestion | ||
| + | More details to come (What, size | quantity | where to get it | price / unit) | ||
| − | |||
Very likely, working in pairs will be ideal for workshops based on these assays, since 'self-reporting' is generally so biased! | Very likely, working in pairs will be ideal for workshops based on these assays, since 'self-reporting' is generally so biased! | ||
(AGiR! is actively investigating this issue, too, since it has effects on both sides of the equation, for producers as well as consumers!) | (AGiR! is actively investigating this issue, too, since it has effects on both sides of the equation, for producers as well as consumers!) | ||
==== Parameters for the Open Source Protocol (still under development!) ==== | ==== Parameters for the Open Source Protocol (still under development!) ==== | ||
| − | ;To get | + | ;To get simplest protocol established, compared with the standard lab techniques |
| − | then To | + | then for each individual: |
| − | : p1 | | + | obtain baseline values (number of micronuclei per thousand cells, or average comet tail lengths/category distributions) |
| − | : p2 | | + | Ideally to get at least 3-5 independent measurements first |
| + | Then, for 'in situ' experiments to vary some lifestyle component, and repeat the assays again. (i.e. exercise, diet, exposures...) | ||
| + | Compare the obtained values - baseline vs. intervention... | ||
| + | |||
| + | ; To measure/ observe | ||
| + | : p1 | number of micronuclei per thousand cells | ||
| + | : p2 | categories of comets (0-5...) | ||
; Constants (Key elements of set-up) | ; Constants (Key elements of set-up) | ||
| − | : c1 | | + | : c1 | |
: c2 | : c2 | ||
| − | |||
| − | |||
| − | |||
| − | + | ;To get Open Source protocol out to the DIT Research communities | |
| − | + | ||
| − | + | Get workshops running, to familiarise people with the technique. Already have interest from Toni B in Melbourne.<br> | |
| − | + | Start gathering acquired baselines for individuals, who will either do ex situ experiments on their own cells, or vary something in their lifestyle (e.g. eat chocolate or blueberries, go running, stop drinking, etc.), and then start collecting 'big data' as more people join in this effort! | |
| + | <br> | ||
| + | |||
| + | === Phase 1 === | ||
| + | To get going with classic methods, on cheek cells, esp following the protocol of Szeto et al (2005) for the comet assays, and [http://wiki.hackuarium.ch/w/Micronucl staining fixed cells for the micronuclei]... | ||
| + | Try proteinase K only treatments (no trypsin) for comet cells to start. Cells simply dissolved away first few times, never seemed feasible to even run the electrophoresis step! | ||
| + | <br> | ||
| + | On the 4th set of attempts (2dec16), by watching carefully as they were being digested, the reaction could be stopped early enough for some visible nucleoids to remain. <br> | ||
| + | Simply running the slides for the electrophoresis step in TBE, allowed the formation of the first cheek cell 'comets' in the Hackuarium! <br> (thanks to Niko at DMF for letting me go on the departmental scope very quickly that afternoon! :) | ||
| + | <br><br> | ||
| + | [[File:CheekCellComet 2dec16slide2x10GFPb.jpg]] This cheek cell 'comet' was imaged by epifluorescence after staining with SYBR-Safe dye.<br><br> | ||
| + | [[File:ControlCheekCells_sybrsafe_2dec16slide1x10bGFP.jpg]] These are control cheek cells that were embedded and treated with all buffers, but never exposed to proteinase K and not run during the electrophoresis step.<br><br> | ||
| + | [[File:H2O2treatedCheekCell 2dec16positivecontrol.jpg]] This 'positive control comet' was obtained after treatment of embedded cheek cells also with hydrogen peroxide, before the proteinase K step. <br> <br> | ||
| + | |||
| + | |||
| − | + | More to come! Who else wants to try?<br> | |
| − | + | update 2017oct14: more on the difficulties in getting to the 'kitchen sink' level, and Anna's 8 weeks as an intern ('faire un stage' is the local term) was still not fully discussed... | |
| + | <br> | ||
| − | == Run / prototype #2== | + | === Run / prototype #2=== |
<!-- Experiment code / name ? --> | <!-- Experiment code / name ? --> | ||
| − | What's new compared to #1: | + | What's new compared to #1: getting the comets to show up with a non-fluorescent staining protocol (if possible)! |
| + | |||
| + | === Run / prototype #3=== | ||
| + | What's new compared to #2: | ||
| + | Development of a microfluidic chip in a slide with miniaturised electrode links, to run the comet assays in a miniformat, that can then be directly examined on a basic microscope... | ||
| + | Design and implementation of this cheek cell comet device may be a key element for a crowd funding campaign (aiming for September 2017). | ||
== Results / Feedback from use == | == Results / Feedback from use == | ||
| + | |||
| + | === Lab Notebook & News (Real) === | ||
| + | Protocols and measurements to be linked from here: [http://wiki.hackuarium.ch/w/AGiR!_for_genomic_integrity/cometassaydev comet protocol development page] | ||
| + | |||
| + | === Foldscope Development for DIT users=== | ||
| + | That this very simple 'origami' Foldscope will allow systematic cell counting and imaging for both micronuclei and comet assays does not seem very likely, after [http://wiki.hackuarium.ch/w/Foldscope many tests].<br> The 140x ball lens, with just a 'sweet spot' in focus makes it quite difficult to score the 1000 cells on a slide we think necessary for quantitation, even though we had thought it might be readily possible for teams of 2; or possibly alone, with further adaptations... | ||
| + | (see youtube videos on Foldscope adaptations for high magnification with a condenser rig, and for fluorescence...)<br> | ||
| + | This is a bit of a disappointment. <br> However, we are how looking into another open microscopy system, the [[OpenFlexure Scope]], for imaging the potential 'cheek cell chips' we hope to develop. | ||
| + | <br> | ||
| + | |||
| + | ===OpenFlexure Scope for Epifluorescence=== | ||
| + | The 3D printed scope developed in England (Bath) by Richard Bowman and colleagues, Julian Stirling and others, may be good for imaging the SybrSafe -stained cheek cell comets, we hope. 3D printing of parts and ordering of many pieces to put it together, including filters became a big push during April 2019. The first 'build' was started 23 May, with the help of Anna D, and Masoud helped start getting the motors together and the software installed in the lab's fedora machine. <br> | ||
| + | <br> The filter set is fitted and signs that the epifluor will work were already obtained with filter paper soaked in fluorescein, but the first 'sybr-safe' -stained cells were not easy to see with the epi-illum... We had more work ahead to get the motor boards to work and the cell imaging, but got there eventually. Stained nuclei were visualised, but comet tails had still not been, even with the hi-power blue LED, with this RaspberryPi camera, before the pandemic era really set in. However, we have some hopes that with a more hi-res camera (now available) we may have more luck in this for the future, also for the 'cheek cell chip' development overall. This would allow panoramas of complete samples of embedded cells, first for scoring micronuclei, then (after appropriate fluidics and a short e-phoresis step and staining) for imaging the comets from the same cells. <br> | ||
| + | Looking forward to more fun ahead! | ||
| + | |||
== Really fun initial citizen science trial - testing chocolate's effects on genomic integrity == | == Really fun initial citizen science trial - testing chocolate's effects on genomic integrity == | ||
| Line 131: | Line 193: | ||
== Recommendation + next steps == | == Recommendation + next steps == | ||
| − | spread the word! | + | Check out the levels of DNA damage in your own cheek cells:<br> |
| + | Here is the [https://www.eventbrite.com/e/workshop-open-sourcing-dna-damage-detection-for-citizen-science-tickets-43050819159 link to the 2018 atelier] (21April2018) in Renens. <br> | ||
| + | |||
| + | Check out the [http://wiki.hackuarium.ch/w/Moss_Menageries_with_AGiR! Moss Menagerie] project also, esp as there is news about how [https://doi.org/10.1016/j.cub.2024.03.019 up-regulating DNA repair] helps [https://elifesciences.org/reviewed-preprints/92621 waterbears resist radiation] challenges - <br> | ||
| + | and<br> | ||
| + | spread the word!<br> | ||
www.genomicintegrity.org | www.genomicintegrity.org | ||
[[Category:Work In Progress]] | [[Category:Work In Progress]] | ||
[[Category:Manual]] | [[Category:Manual]] | ||
Latest revision as of 15:18, 18 September 2024
Genomic integrity is a 'big picture' concept to aid public health efforts and basically includes all the molecular genetic details of cells.
So many things we commonly choose to do can impact genomic integrity, also potentially affecting future generations.
To Note: the concept of 'dynamic genomic integrity' celebrated its 15th anniversary already.
It was first proposed in 2009, as part of the old EFV Prize (to help End Fashion Victimisation)...
(AGiR! as a Swiss Association began over 11 years ago, with the first general assembly meeting held on 16sept2013)
At Hackuarium, AGiR! hopes to get participative research actions really going - developing DIT research methods to readily assay *your very own* cells for DNA damage, using classic (circa 1980s) techniques. If these can already be sold in 'kits,' maybe it will also be simple to make them available to all for open source efforts?
Here is the basic plan: using inner cheek cells for micronuclei and comet assays, because there are still many things we can choose in life, to help protect genomic integrity, even if this is only one way to measure it (actual DNA breaks)!
Inspired by GOSH2018, here also is the AGiR! summary flyer in Chinese
(left), to help everyone think about these issues.
If you want the quick idea, without reading too much more, the poster explaining genomic integrity projects at Hackuarium is worth checking out!
Watch this space for more info and visit the official website of AGiR! Association
AGiR is also carrying out the following projects in the Hackuarium community laboratory:
and, if this inspires you, what do you think about joining in on a new project about swarming bacteria like these?? (potentially!)
Introduction
Context
This is a part of a Hackuarium project by AGiR! on genomic integrity, to find out more and help raise awareness in the public interest.
This wiki page is under construction, and will help keep track as we develop open source simple protocols of two classic methods for quantitation of DNA damage in human cells, the micronuclei and comet assays.
People
Contact and lead:
- Rachel Aronoff - Founder and CSO of AGiR! Action for Genomic integrity through Research!
Participants:
- AGiR! members
- Damian Jurkiewicz
- Tatiana Eliseeva
- Anna Dussuet
- Violaine Regard
- Jennifer Veillard
- Vithooban Thavapalan
- maybe you?!
Main Goals
Genomic integrity is a big picture concept which includes basically all the molecular genetic details of cells (from RNAs to DNAs and the epigenetic marks for proper gene expression), and thus is a topic that can be explored in several ways. For the genomic integrity project at the Hackuarium the focus will be on directly quantifying DNA damage. Comet and micronuclei assays were developed in the 1980s and publications about use of these assays on human cheek cells are already in the literature. These methods, as open source protocols, will encourage citizen science and raise awareness. Additionally, they can provide an easily "workshopable" way to show people more about their own cells, while learning how to assess DNA damage.
In other words, this experiment can be used as a gateway, to demonstrate and raise awareness for everyone on the damage possible from many common substances or activities - on your own cells, your own genetic information!
In the scope of Hackuarium activities, mammalian cell cultures are still (april 2016) a no go, but freshly isolated cheek cells from individuals can be used to do tests on DNA damaging compounds or activities. Such 'ex situ' assays might seem more attractive, especially if potentially toxic compounds are tested! (but the caveat is that it is only after the metabolism of some compounds that the DNA-damaging effects are 'revealed' - for example alcohol to DNA-damaging aldehydes…). For this reason, 'citizen scientists' will be encouraged to first get baseline measures on their cells, then do a 'trial' and test its effects (i.e. daily swimming, chocolate or blueberry consumption, or some other activity). The comet and micronuclei assay results can also be compared after a 'wash-out' stage, without the given trial activity… These would all provide 'biomonitoring' results about the given 'intervention' from baseline conditions.
Another possibility with these assays are to perform ex situ experiments on your own cells, as mentioned, which would be giving acute genotoxicity information...
In order to further investigate compounds that might cause DNA damage, a classic test of bacterial reversion (i.e. the Ames test) or on bulk DNA may also be nice to establish at the Hackuarium.
If enough people follow the 'open source' protocol(s) that we develop to follow controlled exposure protocols or test substances directly on their isolated cheek cells, and clearly significant effects are observed, we may even be able to corroborate or refute previous works in the literature. (Please go to the bottom of the page, for one concrete example being considered.)
Your ideas and comments are welcome throughout btw!
Objectives
- Obtain ready results with cheek cells for the comet and micronuclei assays
- Put together the open source protocols for each method, with suggestions for standard tests and controls
- Demonstrate methods and 'share the love' with other biohacker groups (La Paillasse etc.)
- Study the feasibility of further methods that could contribute to this project - i.e. open-source imaging and microscopy techniques, like the OpenFlexure system, which we are trying for imaging comets by epifluorescence. As of 2019 we had gotten to the point of seeing fluor nuclei, but not comet tails, but then the pandemic caused us to put this to the side, to focus on Corona Detective R&D. In 2022, we hoped to get back to all this 'cheek cell chip' work, and it is still of great interest! You could help!
Cheek cells are continually renewing epithelial cells and are about 50 micrometers in diameter.
Background & Inspiration
Here is the link to the Prezi Rachel presented (13apr16) at an Open hackuarium night! Prezi on DIYbio for Genomic integrity!
Some theory
More on comet cells and micronuclei to come soon!
However, in the meantime, here are the references to the cheek cell papers mentioned in the presentation (and in the prezi):
Szeto et al (2005) Mutation Research 578:371-381
(In this paper, green tea is also shown to be protective.)
dos Santos Rocha et al (2014) Genetics and Molecular Biology, 37(4)702-707
(This paper looks at micronuclei in cheek cells (and also analyses apoptosis) from mouthwash and alcohol users.)
Also here is a new review about micronuclei!
To note: the two assays proposed would be analysing only limited aspects of 'genomic integrity' at the DNA level - large scale chromosomal disruption in the micronuclei assay and double-stranded breaks with the comet assay!
There are many more methods that could look at other aspects (particular nucleotide lesions, reactive oxygen species and inflammation, gene expression, microRNAs, etc.) of genomic integrity!
Inspiration / Similar projects
Inspiration:
- [1] Sylvain Costes and Jon Tang made this great effort in 2014, based upon sub-cellular fluorescence quantification of DNA Damage Response components, but their blood draw kit was a bit tricky for some, and Sylvain told me the results were not so reliable after these fixed cells arrived in the mail, as opposed to their results when they had them 'fresh.'
- [2] This is a Frontiers in Genetics article with a nice focus on using inner cheek cells and other epithelial cells for DNA damage assessment, also from 2014.
- This paper describes results of DNA damage scoring in soy bean farm workers exposed to various pesticides, with DNA damage scored via comet assays of blood cells and micronuclei assays of cheek cells.
Protocol (Plan)
To do both comet cell and micronuclei assays by the simplest possible methods, with the bare minimal laboratory reagents/infrastructure...
To potentially get a 'cheek cell chip' working - so that micronuclei scoring then the comet assay will allow one to assess the level of damage for all the cells in the sample for both methods...
Setup description
General principle for both assays: looking at treated cells on slides through a microscope, imaging and quantitating the effects (i.e. counting number of cells with micronuclei relative to the total number of cells, or determining the length of the 'comet tail' - the longer the 'tail' the more DNA breaks - on average).
The comet assay requires more treatments and a short 'electrophoresis' step, as compared to the micronuclei assay.
For the purpose of cheek cell comet assays, use of a special chamber with mini-electrophoresis capacity, capable of microscopic investigation, is being planned.
Necessary Materials
- Interested citizen scientists
- A toothbrush for gentle collection of the cheek cells
- Lab equipment for the cheek cell 'comet' and micronuclei assays
- Microscope, glass slides
- Lab 'consumables' pipets and tips, beakers
- Agarose, and gel running apparatus for embedding cells, treatments (triton, protease'*, EDTA'**, buffer solutions, etc.) and nucleoid migration (for the comet assay)
- Mercury Lamp Setup and filters for fluorescent dye detection.
- Could also try to design a transilluminator suitable for cellular resolution (UV/Fluo as usual for comets) when imaging (to look for open source models)
- Other reagents and histochemical stains (methylene blue)
- Hydrogen peroxide for positive control cells (oxidative damage)
- Antioxidant molecules as possible 'protective' intervention . (i.e. green tea as in Szeto et al...)
*Proteinase K is recommended in buccal cell protocols, but DIY alternatives are being considered, like tablets for contact lens cleaning
'** To protect the DNA from digestion
More details to come (What, size | quantity | where to get it | price / unit)
Very likely, working in pairs will be ideal for workshops based on these assays, since 'self-reporting' is generally so biased! (AGiR! is actively investigating this issue, too, since it has effects on both sides of the equation, for producers as well as consumers!)
Parameters for the Open Source Protocol (still under development!)
- To get simplest protocol established, compared with the standard lab techniques
then for each individual: obtain baseline values (number of micronuclei per thousand cells, or average comet tail lengths/category distributions) Ideally to get at least 3-5 independent measurements first Then, for 'in situ' experiments to vary some lifestyle component, and repeat the assays again. (i.e. exercise, diet, exposures...) Compare the obtained values - baseline vs. intervention...
- To measure/ observe
- p1 | number of micronuclei per thousand cells
- p2 | categories of comets (0-5...)
- Constants (Key elements of set-up)
- c1 |
- c2
- To get Open Source protocol out to the DIT Research communities
Get workshops running, to familiarise people with the technique. Already have interest from Toni B in Melbourne.
Start gathering acquired baselines for individuals, who will either do ex situ experiments on their own cells, or vary something in their lifestyle (e.g. eat chocolate or blueberries, go running, stop drinking, etc.), and then start collecting 'big data' as more people join in this effort!
Phase 1
To get going with classic methods, on cheek cells, esp following the protocol of Szeto et al (2005) for the comet assays, and staining fixed cells for the micronuclei...
Try proteinase K only treatments (no trypsin) for comet cells to start. Cells simply dissolved away first few times, never seemed feasible to even run the electrophoresis step!
On the 4th set of attempts (2dec16), by watching carefully as they were being digested, the reaction could be stopped early enough for some visible nucleoids to remain.
Simply running the slides for the electrophoresis step in TBE, allowed the formation of the first cheek cell 'comets' in the Hackuarium!
(thanks to Niko at DMF for letting me go on the departmental scope very quickly that afternoon! :)
 This cheek cell 'comet' was imaged by epifluorescence after staining with SYBR-Safe dye.
This cheek cell 'comet' was imaged by epifluorescence after staining with SYBR-Safe dye.
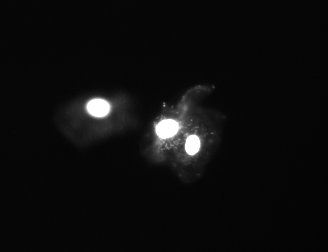 These are control cheek cells that were embedded and treated with all buffers, but never exposed to proteinase K and not run during the electrophoresis step.
These are control cheek cells that were embedded and treated with all buffers, but never exposed to proteinase K and not run during the electrophoresis step.
 This 'positive control comet' was obtained after treatment of embedded cheek cells also with hydrogen peroxide, before the proteinase K step.
This 'positive control comet' was obtained after treatment of embedded cheek cells also with hydrogen peroxide, before the proteinase K step.
More to come! Who else wants to try?
update 2017oct14: more on the difficulties in getting to the 'kitchen sink' level, and Anna's 8 weeks as an intern ('faire un stage' is the local term) was still not fully discussed...
Run / prototype #2
What's new compared to #1: getting the comets to show up with a non-fluorescent staining protocol (if possible)!
Run / prototype #3
What's new compared to #2: Development of a microfluidic chip in a slide with miniaturised electrode links, to run the comet assays in a miniformat, that can then be directly examined on a basic microscope... Design and implementation of this cheek cell comet device may be a key element for a crowd funding campaign (aiming for September 2017).
Results / Feedback from use
Lab Notebook & News (Real)
Protocols and measurements to be linked from here: comet protocol development page
Foldscope Development for DIT users
That this very simple 'origami' Foldscope will allow systematic cell counting and imaging for both micronuclei and comet assays does not seem very likely, after many tests.
The 140x ball lens, with just a 'sweet spot' in focus makes it quite difficult to score the 1000 cells on a slide we think necessary for quantitation, even though we had thought it might be readily possible for teams of 2; or possibly alone, with further adaptations...
(see youtube videos on Foldscope adaptations for high magnification with a condenser rig, and for fluorescence...)
This is a bit of a disappointment.
However, we are how looking into another open microscopy system, the OpenFlexure Scope, for imaging the potential 'cheek cell chips' we hope to develop.
OpenFlexure Scope for Epifluorescence
The 3D printed scope developed in England (Bath) by Richard Bowman and colleagues, Julian Stirling and others, may be good for imaging the SybrSafe -stained cheek cell comets, we hope. 3D printing of parts and ordering of many pieces to put it together, including filters became a big push during April 2019. The first 'build' was started 23 May, with the help of Anna D, and Masoud helped start getting the motors together and the software installed in the lab's fedora machine.
The filter set is fitted and signs that the epifluor will work were already obtained with filter paper soaked in fluorescein, but the first 'sybr-safe' -stained cells were not easy to see with the epi-illum... We had more work ahead to get the motor boards to work and the cell imaging, but got there eventually. Stained nuclei were visualised, but comet tails had still not been, even with the hi-power blue LED, with this RaspberryPi camera, before the pandemic era really set in. However, we have some hopes that with a more hi-res camera (now available) we may have more luck in this for the future, also for the 'cheek cell chip' development overall. This would allow panoramas of complete samples of embedded cells, first for scoring micronuclei, then (after appropriate fluidics and a short e-phoresis step and staining) for imaging the comets from the same cells.
Looking forward to more fun ahead!
Really fun initial citizen science trial - testing chocolate's effects on genomic integrity
As one fun standard protocol for the open source 'citizen science' procedure, which might help resolve controversies in the literature even - for instance, about flavanols in chocolate and what else they might be good for (cardiovascular health, cognition, etc…) - AGiR! is considering proposing to test high and low flavanol chocolate's effects on genomic integrity! This will form one part of the initial citizen science trials for this project! Other potential plans include testing cells in the context of performing activities thought of as 'good' or 'bad' for you, and also testing the cells in isolation (ex situ assays)...
In terms of chocolate: There have been two publications on limited numbers of subjects in terms of flavanols from chocolate protecting skin cells from UV light, based on a sort of 'quantitative skin burn' method - measuring the so-called minimal erythema dose).
Here are the abstracts from two interesting publications in that regard.
http://www.ncbi.nlm.nih.gov/pubmed/19735513
http://www.ncbi.nlm.nih.gov/pubmed/24970388
One of course found a significant effect and the other no significant UV protection of skin. (However a 'Net temple elasticity increased slightly but significantly by 0.09 ± 0.12 mm in the HFC group at 12 weeks compared to 0.02 ± 0.12 mm in the LFC group (MD: 0.06; 95% CI: 0.01 to 0.12 )' and this could be very interesting for many of us! 30g of chocolate a day was the dose, for 12 weeks and a 3 week washout in this study. Have a look! - They even discuss the several reasons that the results are not strictly comparable between the two studies. Trial design is so crucial! )
Maybe in the terms of this genomic integrity project for the Hackuarium, using cheek cells to assess both basal damage levels and damage levels in response to tests of ex situ effects of differing concentrations of hydrogen peroxide, for instance, after eating chocolate with high or low flavanol levels, will be more meaningful than that 'skin burn' model, based on just 30 people. The comet and micronuclei assays are extremely established and clearly quantitative.
Should be fun!
Furthermore, with citizen scientists everywhere involved, we may be able to get much bigger numbers than for those two previous works.
In the meantime…
A collaboration with a very keen 'chocolatier' from Gruyere may help us!
We will try to see which of his dark chocolates contains the most flavanols (and perhaps he will modify some steps in the production process to increase the levels found!) and had a 'degustation' of his grand cru chocolates yesterday! (25 may 2016)
If you are interested in more information and a closer look at the results (in a nutshell, no significant preference was found for the four different chocolates provided, but there was a slight trend leading to two of the four seeming to be more preferred! Additionally, eating any of these magnificent chocolates made a commercial brand less attractive for many participants!), here is a page with further info about this: chocolate tasting 2016
As we are located in the land of chocolate - Switzerland - what could be better? Hoping people will like the idea! For us all!
Recommendation + next steps
Check out the levels of DNA damage in your own cheek cells:
Here is the link to the 2018 atelier (21April2018) in Renens.
Check out the Moss Menagerie project also, esp as there is news about how up-regulating DNA repair helps waterbears resist radiation challenges -
and
spread the word!
www.genomicintegrity.org