Difference between revisions of "QualityControl4Genes"
| Line 9: | Line 9: | ||
Interested to learn more? | Interested to learn more? | ||
| − | Here is part of [https://doi.org/10.7554/eLife.33292 an open access paper]'s introduction more focused on another quality control mechanism - but it all starts with mRNA | + | Here is part of [https://doi.org/10.7554/eLife.33292 an open access paper]'s introduction more focused on another quality control mechanism - but it all starts with mRNA surveillance (or nonsense-mediated decay): |
Nonsense-mediated decay (NMD) (reviewed in [He and Jacobson, 2015]) is a translational surveillance pathway to mitigate deleterious products of premature stop codons. In NMD, recognition of an early stop codon destabilizes an mRNA (Morse and Yanofsky, 1969; Baserga and Benz, 1988; Losson and Lacroute, 1979). Foundational studies in ''S. cerevisiae'' and ''C. elegans'' revealed protein factors responsible for NMD (Leeds et al., 1991; Hodgkin et al., 1989; Pulak and Anderson, 1993). In the decades since, a large body of literature has highlighted similarities and differences in NMD between yeast and metazoans. For example, while both yeast and metazoan NMD involve a core set of three proteins (UPF1-3 in yeast, SMG-2–4 in metazoans), metazoans require additional proteins for NMD (e.g. SMG-1, –5, and −6). Additionally, ''Saccharomyces cerevisiae'' NMD is thought to occur predominantly through decapping and 5’>3’ exonucleolytic degradation (Muhlrad and Parker, 1994), while studies across metazoans have implicated both exo- and endonucleolytic machineries (e.g. [Lykke-Andersen, 2002; Lejeune et al., 2003; Gatfield and Izaurralde, 2004; Glavan et al., 2006; Huntzinger et al., 2008; Eberle et al., 2009; Lykke-Andersen et al., 2014; Schmidt et al., 2015; Ottens et al., 2017]). | Nonsense-mediated decay (NMD) (reviewed in [He and Jacobson, 2015]) is a translational surveillance pathway to mitigate deleterious products of premature stop codons. In NMD, recognition of an early stop codon destabilizes an mRNA (Morse and Yanofsky, 1969; Baserga and Benz, 1988; Losson and Lacroute, 1979). Foundational studies in ''S. cerevisiae'' and ''C. elegans'' revealed protein factors responsible for NMD (Leeds et al., 1991; Hodgkin et al., 1989; Pulak and Anderson, 1993). In the decades since, a large body of literature has highlighted similarities and differences in NMD between yeast and metazoans. For example, while both yeast and metazoan NMD involve a core set of three proteins (UPF1-3 in yeast, SMG-2–4 in metazoans), metazoans require additional proteins for NMD (e.g. SMG-1, –5, and −6). Additionally, ''Saccharomyces cerevisiae'' NMD is thought to occur predominantly through decapping and 5’>3’ exonucleolytic degradation (Muhlrad and Parker, 1994), while studies across metazoans have implicated both exo- and endonucleolytic machineries (e.g. [Lykke-Andersen, 2002; Lejeune et al., 2003; Gatfield and Izaurralde, 2004; Glavan et al., 2006; Huntzinger et al., 2008; Eberle et al., 2009; Lykke-Andersen et al., 2014; Schmidt et al., 2015; Ottens et al., 2017]). | ||
| Line 24: | Line 24: | ||
Here is the [https://drive.google.com/file/d/1Vw_f71P4g2F9_E1V4VNZ4x0B5i1MvB78/view?usp=share_link original paper about the cloned worm gene], about which we hope to learn more. | Here is the [https://drive.google.com/file/d/1Vw_f71P4g2F9_E1V4VNZ4x0B5i1MvB78/view?usp=share_link original paper about the cloned worm gene], about which we hope to learn more. | ||
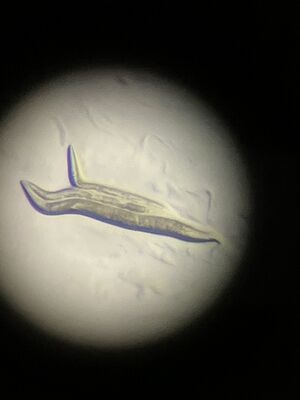
| − | In humans, there are two copies on different chromosomes and more... | + | We have already confirmed old results here, with rescue from PCR products, as shown in the image... |
| + | |||
| + | Sequence confirmation on an old deletion in the gene (but much after the original paper) gives us clues about what is most important for function, and we speculate some essential function, partly because mutations in the gene are rare (and the only point mutations found were on either end of the same intron...). | ||
| + | |||
| + | In humans, there are two copies on different chromosomes, with the X-linked one tied to Intellectual Disabilities, and [https://www.cmmc-uni-koeln.de/de/research/research-areas-projects/research-area-c/niels-gehring-c-07 more interesting features] in knockdown and attempts to knock this QC mechanism out in cells... | ||
| + | |||
Revision as of 19:58, 30 November 2022
Even gene expression needs quality control!
With colleagues in Basel and the approval of of the Federal authorities in Bern, we are looking further into some molecular mysteries around all this...
Our first transgenic animals (well, as an invertebrate some say we should just call them GMO) are in the lab, and we plan to learn a lot by making some more!
Interested to learn more?
Here is part of an open access paper's introduction more focused on another quality control mechanism - but it all starts with mRNA surveillance (or nonsense-mediated decay):
Nonsense-mediated decay (NMD) (reviewed in [He and Jacobson, 2015]) is a translational surveillance pathway to mitigate deleterious products of premature stop codons. In NMD, recognition of an early stop codon destabilizes an mRNA (Morse and Yanofsky, 1969; Baserga and Benz, 1988; Losson and Lacroute, 1979). Foundational studies in S. cerevisiae and C. elegans revealed protein factors responsible for NMD (Leeds et al., 1991; Hodgkin et al., 1989; Pulak and Anderson, 1993). In the decades since, a large body of literature has highlighted similarities and differences in NMD between yeast and metazoans. For example, while both yeast and metazoan NMD involve a core set of three proteins (UPF1-3 in yeast, SMG-2–4 in metazoans), metazoans require additional proteins for NMD (e.g. SMG-1, –5, and −6). Additionally, Saccharomyces cerevisiae NMD is thought to occur predominantly through decapping and 5’>3’ exonucleolytic degradation (Muhlrad and Parker, 1994), while studies across metazoans have implicated both exo- and endonucleolytic machineries (e.g. [Lykke-Andersen, 2002; Lejeune et al., 2003; Gatfield and Izaurralde, 2004; Glavan et al., 2006; Huntzinger et al., 2008; Eberle et al., 2009; Lykke-Andersen et al., 2014; Schmidt et al., 2015; Ottens et al., 2017]).
Although protective under many circumstances, the NMD pathway also contributes to pathological suppression of expression from numerous disease-causing mutations (about 11% of point mutations responsible for human disease [Mort et al., 2008]).
Have questions?
Would you be interested to try some worm research?
Both a Crispr knock-out and knock-in (mCherry) and RNAi are planned, too!
Here is the original paper about the cloned worm gene, about which we hope to learn more.
We have already confirmed old results here, with rescue from PCR products, as shown in the image...
Sequence confirmation on an old deletion in the gene (but much after the original paper) gives us clues about what is most important for function, and we speculate some essential function, partly because mutations in the gene are rare (and the only point mutations found were on either end of the same intron...).
In humans, there are two copies on different chromosomes, with the X-linked one tied to Intellectual Disabilities, and more interesting features in knockdown and attempts to knock this QC mechanism out in cells...
contact rachel (at) hackuarium (dot) ch...
=)