Difference between revisions of "MCBP3"
| (One intermediate revision by the same user not shown) | |||
| Line 32: | Line 32: | ||
<br> | <br> | ||
| − | and here | + | and here is the [http://wiki.hackuarium.ch/w/CBEMresults the page summarising] sampling for the project, which began in earnest for the second summer on 12 June 2017! <br> |
| − | + | Hope you were able to join us! :)<br> | |
<br> | <br> | ||
| Line 60: | Line 60: | ||
This is not really in progress now (2020 - but we are having a last control summer sampling set... NoJazz!)! <br> | This is not really in progress now (2020 - but we are having a last control summer sampling set... NoJazz!)! <br> | ||
All the [https://kc.kobotoolbox.org/hammerdirt/reports/Microbio_2017/export.html data were already available on line] in the kobo site and Roger made great headway on the python coding in [https://github.com/hammerdirt/water-quality-2016-2017 github]. <br> | All the [https://kc.kobotoolbox.org/hammerdirt/reports/Microbio_2017/export.html data were already available on line] in the kobo site and Roger made great headway on the python coding in [https://github.com/hammerdirt/water-quality-2016-2017 github]. <br> | ||
| − | Just to give you a little idea, here is something | + | Just to give you a little idea, here is something put together in 2017, only looking at average UV+ colonies (thus 'confirmed' ''E. coli'', our bioindicator for dirty, untreated sewage water).<br> |
[[http://wiki.hackuarium.ch/images/9/9e/GraphUV%2B.pdf]] | [[http://wiki.hackuarium.ch/images/9/9e/GraphUV%2B.pdf]] | ||
Latest revision as of 13:12, 12 July 2020
The Montreux Clean Beach Project Microbio team has been looking forward to replicating last summer's water sampling (for DIT research!) and is beginning again at Hackuarium:
(If someone would like to help for translations to French or other languages, please do feel free!)
Inspiration
Many EasyGel plate results have already been obtained, from last year, and this year we will have fluor confirmation for our E. coli colonies!
A new sort of card from Micrology labs is also being tested!
First Preliminary Controls at Hackuarium
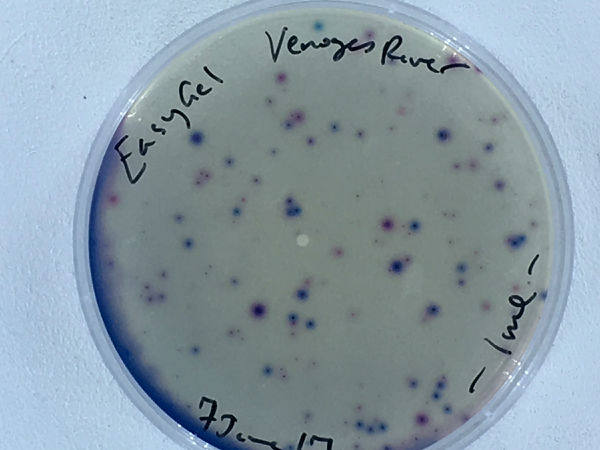
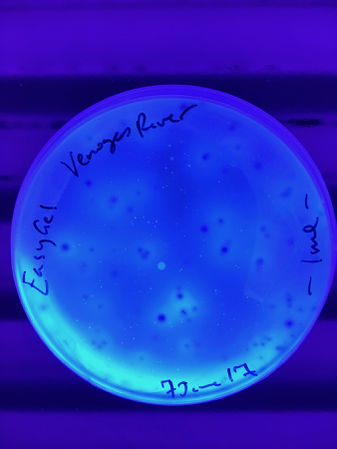
The Venoge River was sampled on 5June for a single plate and card test, which are shown in the Gallery below.
Key Timing Criteria
We want to make sure to get good images of the colony forming units (CFUs) at 24h, when the fluorescence is not too dispersed, and at 48h, when the chromogenic media results are at their peak.
Watch this space for more soon!
Gallery
The first images are 24h images of the first EasyGel plate, bright field and UV fluor, and the first ECC card test ...
and here is the the page summarising sampling for the project, which began in earnest for the second summer on 12 June 2017!
Hope you were able to join us! :)
Timeline for Sampling
8 weeks starting 12 June 2017
Every Monday until 31 July 2017
Imaging and Scoring Tuesdays for good fluor comparisons to bright field, and Wednesdays for 48h chromogenic results.
(so through until 2 August for initial scoring)
As for last year, samples must be kept cool and plated within 5 hours of their collection.
Labelling and note-taking will be key, and any deviations from this must be noted carefully!
Data collection and analyses will also be facilitated by IT tools, to be described more fully in a further section of this page. (RE)
Montreux Bay Sites: Montreux Rive Droit, Sauvetage, Vernex
Control Sampling: River water (Venoges or Vuachère Rivers) for positive controls; Tap water for negative control.
Data Analyses and Reporting process
This is not really in progress now (2020 - but we are having a last control summer sampling set... NoJazz!)!
All the data were already available on line in the kobo site and Roger made great headway on the python coding in github.
Just to give you a little idea, here is something put together in 2017, only looking at average UV+ colonies (thus 'confirmed' E. coli, our bioindicator for dirty, untreated sewage water).
[[1]]
Lake water has much more of such bacteria than the river water (our standard 'positive control'), but a clear peak at unhealthy levels (about 200 E.coli per 100ml is not super horrible, but it is not healthy, and a clear increase over the very low baseline!) was found around the time of the jazz, and this does not seem to be simply explained by rainfall.
Further information
- The original project page from last summer's sampling
- The hammerdirt site - with cleaning up local beaches as data collection exercises!
- The Micrology Labs list of media, including ECA Check Plus EasyGel plates.
- website - Action for Genomic Integrity through Research!