Difference between revisions of "Micro to Macro Water Pollution"
Hammer dirt (talk | contribs) |
Hammer dirt (talk | contribs) |
||
| Line 320: | Line 320: | ||
[[file:Grownplates.jpg|thumb|400px|right| Vidy and Vauchère 24hours methylene blue agar, June 2016.]] | [[file:Grownplates.jpg|thumb|400px|right| Vidy and Vauchère 24hours methylene blue agar, June 2016.]] | ||
[[file:Upclose2.png |thumb|400px|right|E.coli colony Vauchère methylene blue agar, June 2016.]] | [[file:Upclose2.png |thumb|400px|right|E.coli colony Vauchère methylene blue agar, June 2016.]] | ||
| − | The samples were removed from the incubator after 24hours of incubation at 37c. Mulitple colonies were noted in the methlylene blue agar, | + | The samples were removed from the incubator after 24hours of incubation at 37c. Mulitple colonies were noted in the methlylene blue agar, we are displaying only the counts of the ''E.coli'' colony forming units. |
<br> | <br> | ||
There was no visible colony forming units in the tap water or the sample taken from Vidy (the unknown). The colony forming units of ''E.coli'' counted from the Vauchère samples are presented in the table below. | There was no visible colony forming units in the tap water or the sample taken from Vidy (the unknown). The colony forming units of ''E.coli'' counted from the Vauchère samples are presented in the table below. | ||
| Line 349: | Line 349: | ||
<br> | <br> | ||
<br> | <br> | ||
| − | The ECA Check Easygel material arrived on June 11th so | + | The ECA Check Easygel material arrived on June 11th so as a control that they work as expected - 0.5ml of the same Tap Water and Vuachère (saved in the refrigerator) samples were plated. The EasyGel is much easier to distinguish the colony forming units than the methylene blue agar. |
<br> | <br> | ||
[[file:Easygelplates.jpg |thumb|500px|none|ECA Check colonies at 24 hours, June 2016.]] | [[file:Easygelplates.jpg |thumb|500px|none|ECA Check colonies at 24 hours, June 2016.]] | ||
Revision as of 15:38, 13 June 2016
Community based environmental monitoring has been proven effective in a variety of fields, water quality monitoring by volunteers is in wide practice across North America. Four Swiss nonprofit associations use visible pollution inventories, microbiological monitoring and chemical analysis by combining classic laboratory techniques and the use of innovative bio-reporters to expand the capabilities of community based environmental monitoring.
Aims
To effectively integrate more advanced and innovative water quality monitoring techniques to classic community based environmental monitoring activites.
Collaboration Defined
| Partner | Role |
|---|---|
| ACTION FOR GENOMIC INTEGRITY THROUGH RESEARCH! | Technical oversight for the microbial assays, training of hammerdirt staff, quality control of laboratory operations and of sample processing and analyses. |
| HACKUARIUM | Laboratory infrastructure and security, testing equipment and supplies for biologicals |
| HAMMERDIRT | Collection and processing of samples, data storage and administration. Communication of analyses and results. |
| BIODESIGN.CC | Analysis of samples for mercury, Cadmium Zinc, Alkanes and arsenic using the bio-reporters |
Water Quality and Analysis
Spring and Summer 2016
Introduction
One of the benefits of a “Community Based Environmental Monitoring” (CBEM) project is to expand the monitoring capacity of government agencies and engage the public in environmental stewardship. In response to questions from the public and our own professional curiosity we have decided to initiate a limited CBEM project (1) (2) .
As part of the Montreux Clean Beach Project II(MCBPII), a water quality testing (WQT) program is being initiated at selected sites of the MCBPII. Symptoms reported by members and frequent bathers in the lake during previous summer seasons have sometimes been attributed to low bathing water quality, influenced by increased anthropogenic pressure of lakefront activities during certain times of the year. This year's WQT aims to gather microbiological data over a six week period in order to test this hypothesis.
Meetings at “La Maison de la Rivière” and the “Institut National de Recherche Agronomique” support the idea that the WQT information proposed is not regularly collected at the Haut-Lac locations covered by the MCBPII.
Objectives
CBEM programs are designed to educate the public, involve local citizens in environmental stewardship and provide reliable data for government and other interested organizations. In this regard the MCBPII CBEM program is no different than that of any other existing project. (2)(3)(4)(5)(6)
Specifically, the goals for this project are to:
- Collect reliable water quality data as defined by the World Water Monitoring Challenge
- Collect and enumerate the quantity of key bacteriological indicators, in particular those related to fecal contamination
- Increase the skills of hammerdirt staff in regards to collecting and processing water samples.
- Make the obtained data free and open to use for all concerned
- Inform the public about affordable and simple water monitoring techniques
- Inform the public of community based resources designed to advance scientific literacy
More information CBEM Network
Deliverables
The objectives of a CBEM are designed to increase the monitoring capacity of current public systems and provide reliable, accessible data for the public and other government researchers. Data gathered from this project will be reported with our regular activity reports and the raw data will be available for download within 72 hours of the testing date from our repository on Github or through the KoboToolbox for the location in question. This document along with sampling protocols and all other supporting documents will be available on the wiki.
Weekly results will be furnished to the CIPEL, SIGE and INRA:
- Location and number of tests
- Results of tests completed for the week
An end of project report will be published in PDF format, available for interested parties, and include:
- Summary statistics and findings
- Graphs/plots
- Map overlay with densities and values
- Comparison with government supplied data (if available)
- Discussion of limitations and possible improvements to future CBEM projects
More information KoboToolbox
Testing Materials
Heavy Metals
Bioreporters, designed and fabricated by biodesign.cc will be used to test for Alkanes, Arsenic, Mercury and Cadmium/Zinc.
What are bioreporters ? Get some background information here
Turbidity, temp, DO and PH
Testing materials and supplies will be procured from the World Water Monitoring Challenge an international program managed by the International Water Association and the Water Environment Federation.
Assembled and sold by Lamotte company as a “basic test kit,” each kit includes one set of hardware and enough reagents to conduct up to 50 rounds of testing for pH, dissolved oxygen, temperature, and turbidity. Included are:
- Instruction booklet
- Sample collection jar
- PH test tube
- Dissolved oxygen vial
- Secchi disk decal
- Temperature strips (14-40°C and 0-12°C)
- 50 pH reagent tablets (enough for 50 tests)
- 100 Dissolved oxygen reagent tablets (enough for 50 tests)
- Color chart for determining DO, pH and turbidity test results
Microbiological Analyses
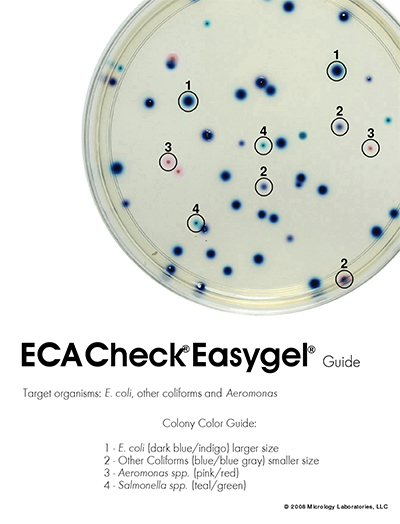
Presence and quantity of E.coli, other coli forms, Salmonella and Aeromonas will be tested using ECA Check Easygel. Easygel was first approved for Water Watch volunteer monitoring programs in the USA in 1999. A study released in 2009 compared the results of “Easygel” and “3M Petrifilm” used by volunteers to the results of laboratory analysis. Both methods had an overall accuracy rate above 80%. One big benefit of the Easygel format is that environmental samples of up to 5ml can be readily plated, with no worries about having to melt (and cool down!) agarose as used in standard microbial plates.(7)(8)(10)(14)
Members of the Hackuarium laboratory have used “Easygel” in previous educational activities, thus facilitating the training of hammedirt staff and volunteers.
Sampling Methods
Water samples will be taken just below the surface (0.5m) and collected in sterile containers with a secure, air-tight lid.
Advantage: simple and inexpensive, an important consideration to a program on a budget. It also does not require any special sampling apparatus. The volunteer holds a sampling container under the surface of the water, expells visible air bubbles, and closes the container under the water (to avoids losing dissolved gasses, particularly important if testing for volatile analytes).
Disadvantage: vertical gradients of temperature, oxygen, nutrients, and algae may be missed in these near surface samples. Deep lakes have a thermal gradient in the summer, and Lac Leman has at least two thermal regions (and, possibly, at least two chemical regions as well). Even in shallow, usually well-mixed lakes, chemical gradients will develop during calm periods.
Sampling methods by other CBEM projects : Stepenuck et all PDF, Fact Sheet Volunteer Water Monitoring, SOP Moore Creek E.coli sampling PDF, Texas Sampling PDF
Heavy Metals
Two samples of 250ml will be taken and transported to the hackuarium for later analysis. One sample will be acidified to fix the heavy metals in solution.
Turbidity, temp, DO and PH
One sample will be taken using the container supplied with the testing kit and processed immediately.
Microbiological samples
Three independent samples will be taken from each designated site in 15ml conical tubes pre-labeled in the following manner:
- Location
- Date
- Sample number for that day and location
- Time of sample
Once collected, the samples will be placed on ice and transported to the facility at Renens for plating on the ECA Check plates within 6 hours. Label information will be recorded in the field notes and the “KoboToolBox” for that sampling date and location.
Processing samples and recording results
Turbidity, temp, DO and PH
The instructions provided with the kit will be followed and the results will be noted in the field manual and entered into the “KoboToolbox” form for that sampling date and location.
Microbiologicals
One petri dish per sample will be labeled in the same manner as the sample container with the addition of the quantity of sample added to the media. A 1-5ml of sample water will be mixed into the provided gel media solution, plated and incubated for about 18 hours at the Hackuarium facility in Renens.
After incubation, plates will be scored, counting total colony forming units (CFU), and each identifiable species (E. coli CFUs will appear dark blue, other coliform CFUs will appear lighter blue-gray or blue-violet, Aeromonas spp. will appear as pink to very light pink, and Salmonella spp. will appear as light green colonies). Data will be reported as CFU/100ml of sample.
With values from three independent samples at each site, means and standard errors can be used to test significance of the obtained data over the course of the sampling period.
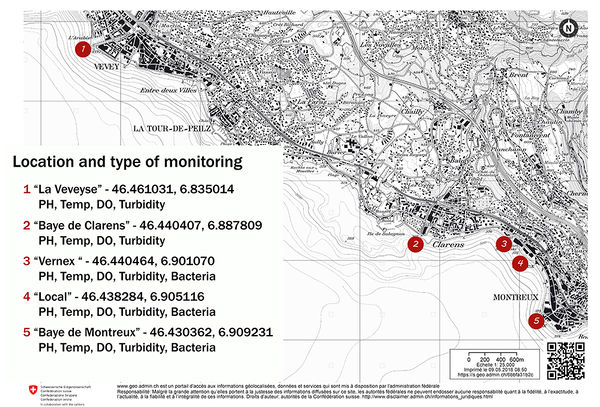
Locations
Three of the five locations are currently part of the MCBPII beach-litter-survey project. Point 4 will be added to the beach litter survey by using a net to catch visible pollutants on the water surface below the boat dock. Point 3 will only be monitored for chemistry and biologicals. Correlations of the test results with the physical location of the sampling will be of great interest.
To see a map of the locations that benefit from regular water quality monitoring : Eau de baignade
Testing Schedule
Calendar from biodesign.cc to be embedded here
key dates to be defined :
- #Qualification Day June 7, 2016 "Q-Day"
- Event public demonstration of sampling and plating
- Start date for chemical samples
- Start date for microbiologicals
- Event public discussion of results, improvements and future projects
Microbiologicals
Testing will begin about 20 June 2016, and weekly samples will be taken from locations 3, 4 and 5. A 'negative control' sample will also be obtained each week from a local drinking fountain (labelled 'eau potable'). A positive control from an urban river (like the Vuachère or other) is also possible.
If a Tuesday afternoon testing protocol is followed over 7 weeks, the timeline would run from 21 June -2 August, require 21 sterile 15ml conical tubes for the water sampling, and 21 ECA Check EasyGel plates per site. After including a single 'negative control' for each sampling day, this will require 70 plates.
In case there are too many bacteria to count (TMTC) from certain samples, samples will be saved at 4oC and further dilutions can be also plated and scored.
Turbidity, temp, DO and PH
Testing will begin on the Tuesday or Thursday the week the kits are received (late May). The kit contains enough reagents to conduct 50 tests, thus allowing us to monitor the locations for 10 weeks. Data will be carefully recorded, digitized and analyzed.
Field Blanks
Field blanks are used to assess potential contamination from sample handling, airborne materials, equipment, media, and other sources. A field blank usually consists of a sterile diluent sample of 2 mL that is taken to the site and poured into a properly labeled sample container during the first bacteria sampling event of that day. The blank sample is collected in the same type of container, labeled as a field blank, and handled and analyzed along with all the bacteria samples collected on that day. It is used to identify errors or contamination in sample collection and analysis.
Data
Handling and storage
The addition of water quality testing will require changes in our digital form and reporting methods. The form currently used by surveyors will change, thus separating the data in two sections, one section prior to WQT and one after. Data for the two sections will have to be merged prior to any analysis. The results of bacterial testing will be available 48-72 hours after sampling, thus adding another step to the survey. This should not, however, change the availability of the other survey results which is currently 24 hours after each survey.
Data analysis and presentation
The existing data set for MCBPII includes visible pollution surveys, hydrologic and meteorological data. With the addition of microbiological and basic water chemistry a more complete picture of the environmental status of the locations in question can be established.
Two interpretations of the microbiological data will be used: the geometric mean and the 95th percentile. The geometric mean is in common use in North America and the 95th percentile method conforms to EU directive 2006/7/EC. Furthermore, any publicly available water quality data in the area will be presented and integrated where appropriate.
The relationship between the results of WQT and the quantity and type of visible pollutants will be explored.
How is the water currently monitored and evaluated ? Here is the source document from the Swiss Confederation Eaux de baignade
References
Sampling and Analysis in Community Based Environmental Monitoring
- Assessing the performance of volunteers in monitoring Streams. Fore, Leska, Paulsen, Kit and O'Laughlin, Kate. Bellvue, WA : Blackwell Science, 2001, Fresh Water Biology. Fore et all pdf
- Volunteer environmental monitoring and the role of the universities : The case of Citizens Environment Watch. Savan, Beth, Morgan, Alexis and Gore, Chritopher. Toronto : s.n., June 2003, Environmental Management, Vol. 31, pp. 561-568. Savan et all PDF
- Nicholson, Emily, Ryan, Jane and Hodgkins, David. Community Data – where does the value lie? Assessing confidence limits of community collected water quality data. Victoria : Waterwatch Victoria c/o Department of Natural Resources & Environment, 2002. Nicholson et all PDF
- A review of citizen science and community-based environmental monitoring: issues and opportunities. Conrad, Cathy and Hilchey, Krista. [ed.] Springer. Halifax, Nova Scotia : s.n., 2010, Environ Monit Assess, pp. 273-291. Conrad & Hilchey PDF
- World water monitoring challenge. Instructions. Geneva : World water montioring Challenge, 2015. Instructions for the water monitoring test kit provided by the world water monitoring challenge. Instructions for PH, DO, temp and turbidity PDF
- World Health Organization. Guidelines for safe recreational water environments. Geneva : World HEalth Organization, 2003. Vol. 1 Coastal and Fresh Waters. WHO Guide Lines
- Texas Stream Team Volunteer Water Quality Monitoring. E.coli Monitoring and Analysis Procedures. 2010. Texas Sampling PDF
- Stepenuk, Kristine, et al. Volunteer monitoring of E. coli in streams of the upper Midwestern United States: a comparison of methods. [ed.] University of Wisconsin—Extension. Madison : Springer, 2010. Stepenuck et all PDF
- Myers, D.N., et al. Fecal indicator bacteria (ver. 2.1). [book auth.] U.S. Geological survey. U.S. Geological Survey Techniques of Water-Resources Investigations. 2014, Vol. book 9, chap. A7, section 7.1. Myers et all PDF
- Iowa Department of Natural Resources, Geological Survey. Citizens Monitoring Bacteria. Iowa City : s.n., 2005. Vols. Water Fact Sheet 2005-1.Fact Sheet Volunteer Water Monitoring
- Arizona Department of Environmental Quality Water Quality Improvement Grant Program. Writing Effective Monitoring Plans. s.l. : Arizona department of environmental quality, 2008. TM 08-04. Guide to writing a sampling plan PDF
- USEPA REGION 9 LABORATORY. STANDARD OPERATING PROCEDURE FOR VOLUNTEER MONITORING OF SURFACE WATERS FOR BACTERIA. Richmond : United States Environmental Agency, 2007. SOP Volunteer monitoring PDF
- U.S. Geological Survey. National Field Manual for the collection of water quality data. 2006. Vol. Book 9. http://water.usgs.gov/owq/FieldManual/
- Wolfson, Dr. Lois. E. coli Monitoring – Effective Techniques and Test Kits for Volunteers. Michigan Clean Water Corps Conference. s.l. : Michigan State University, October 16, 2007. Test Kit Presentation PDF
- Epstein, Charlotte. KNOWLEDGE AND POWER IN GLOBAL ENVIRONMENTAL ACTIVISM. s.l. : International Journal of Peace Studies, 2005. Epstein PDF
- CH. Loi fédérale sur la protection des eaux. [Online] juin 1, 2014. https://www.admin.ch/opc/fr/classified-compilation/19910022/index.html#a4.
- Kleehammer, Katie and Sigler, Adam. Moore Creek Volunteer Monitoring for Escherichia coli - Sampling and analysis. Bozeman : MSU Extension Water Quality, 2012. SOP Moore Creek E.coli sampling PDF
- Homogeneous Distribution of Escherichia coli Measured within the Vertical Water Column of Small, Freshwater Streams. Buckalew, David, et al. Farmville, VA : Scientific Research Publishing, March 24, 2014, Water Resource and Protection, pp. 410-421. Buckalew et all PDF
- Micrology LAboratories. Easy Gel Method Procedure. Goshen, Indiana : s.n., 2015. Recomended procedures for chromogeinc media. Procedure Easygel PDF
- Micrology Laboratories. ECA Check Easy guide. Goshen : Manufacturers color guide for chromogenic media. ECA guide to determining bacteria PDF
- DIRECTIVE 2006/7/EC concerning the management of bathing water quality. European Union. Strassbourg : s.n., 2006, Official Journal of the European Union. Directive 2006/7/EC
- Canfield, Daniel; Hoyer, Mark; Volunteer Lake Monitoring : Testing the Reliability of Data Collected by the Florida LakeWatch Program: Lake and Reservoir Management-March 2002 Reliability of volunteer data PDF
Documents from hammerdirt Clean Beach Projects
- Activity report Nov 2015 - April 2016
- Pilot project CBEM May 2016
- Hammerdirt Clean Beach Project 2 2015 Draft
- Comparison of Lake Geneva beaches to other OSPAR regions
- Citizen Science and litter studies (french)
- MCBPI 2014-2015 results (French)
- MCBPI 2014-2015 results (English)
Qualification Day
As indicated in the project plan hammerdirt volunteers met with the technical advisor from AGIR and agreed to hold a water sampling and sample handling qualification day (Q-Day) prior to the beginning of the Water Quality Testing (WQT) pilot project.
Objectives of Q-Day
- Combine a litter survey with the collection of water samples
- Collect samples in accordance with the protocol for the WQT project
- Prepare culture medium using available laboratory equipment
- Collect samples without introducing contaminants
- Prepare samples without introducing contaminants
- Execute all tasks following laboratory safety regulations
- Verify that all laboratory equipment is operational
Meeting objectives
The success of Q-Day was based on the response to the following questions:
- Were the samples collected and plated within a 6-hour window?
- Was the litter inventory completed prior to leaving the site?
- Did field team observe proper hygiene and sampling protocols?
- Did all laboratory equipment function? Were the team members able to use equipment?
- Did the negative controls show any signs of growth?
- Did the positive controls show growth in accordance with historical results?
The survey sites for Q-Day
- Positive control Vauchère river
- Negative control : tap water
- Unknown : Bay de Vidy
Combining a litter survey with water sampling
A comprehensive litter survey can take up to two hours (depending on the size of the beach). For the Q-day exercise hammerdirt chose a small beach in an urban center. Two risks were immediately identified:
- Cross contamination of samples from handling litter and water sampling
- Running out of time within a 6-hour window to plate the samples
It was decided that initially the litter would be collected at the site by all team members and then, at the discretion of the team members, one individual would cease handling litter and collect the required samples.
11:50 Litter collection and inventory
12:50 Sample collection
13:40 Arrival at the laboratory
Collect samples in accordance with the protocol
The location was easy to access. The team packed soap, hand sanitizer and disposable gloves specifically for water sampling operations. The designated individual ceased handling litter and washed hands with soap and water. Clean, disposable gloves were used for the sampling process. Samples were collected by wading into the water and plunging the sampling tube roughly 50cm deep. The sampling tube was uncapped underwater and filled completely. Once the sampling tube was out of the water a portion was poured off and the sampling tube was capped and put on ice. All team members washed hands and used hand sanitizer at the end of the operation.
Prepare culture medium using available laboratory equipment
For Q-Day operations we used a dehydrated culture media, eosin methylene blue agar, produced by Oxoid and sold under product number CM0069. The culture media that was ordered and that will be used for the WQT project, ECA Check Easygel, had not arrived by the scheduled date. The methylene blue agar requires significantly more preparation and handling than the ECA Check Easygel. The following steps needed to be completed prior to plating (within the 6-hour limit):
- Suspend 37.5g in 1 litre of distilled water.
- Bring to a boil to dissolve completely.
- Sterilise by autoclaving at 121°C for 15 minutes.
- Cool to 60°C
- Shake the medium in order to oxidise the methylene blue
- Pour medium into sterile petri dishes
- Cool medium to solid and introduce water samples
- Prepare samples without introducing contaminants
Collected samples were stored in the laboratory specimen refrigerator at 4c while the chromogenic medium was prepared. The work surface, a fume hood, was sterilized with 70% ethanol prior to pouring the chromogenic media into disposable, sterile petri dishes. The surface was sterilized again prior to plating of the water samples.
Sterile, disposable pipettes were used and changed for each water sample. Participants used disposable latex gloves and a Bunsen burner was lit during all pouring and plating operations.
Three samples from the positive control and the unknown were plated as well as one sample from the negative control (tap water).
Plated samples were covered and placed in the incubator at 37c for 24hours.
Were the objectives met?
- Were the samples collected and plated within a 6-hour window? Yes, samples were collected at 12:50 and plated at 18:30.
- Was the litter inventory completed prior to leaving the site? Yes the results can be seen here :Vidy inventory
- Did field team observe proper hygiene and sampling protocols? Yes, field participants prepared and utilized a field hygiene kit including soap, hand sanitizer and disposable gloves. The depth of water sample was observed.
- Did all laboratory equipment function? Were the team members able to use equipment? Yes, although prior organization and identification of the proper functioning equipment could have reduced the preparation time.
- Did the negative controls show any signs of growth? No, the negative controls exhibited no growth. Indicating that no contamination occurred during sample handling and processing.
- Did the positive controls show growth in accordance with historical results? Yes, in accordance with the experience of the technical advisor.
Remarks
- Field staff will need to organize activities appropriately especially in regards to the times required to inventory litter and the 6-hour time limit for water samples.
- A bottle of water should be included in the field hygiene kit to rinse hands.
- A dedicated space needs to be attributed in the lab for this project.
- The incubator needs to be reserved ahead of time for sample processing (avoid conflicts with other projects).
- The level of the fume hood should be verified, the cooled plates showed a slight unequal distribution.
- A training session with ECA Check Easygel should be scheduled.
Conclusion
An opening day can be scheduled, hammerdirt staff are competent and all the necessary equipment is available at the lab.
Results of Microbiological Analyses
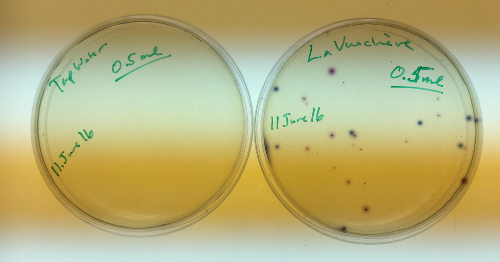
The samples were removed from the incubator after 24hours of incubation at 37c. Mulitple colonies were noted in the methlylene blue agar, we are displaying only the counts of the E.coli colony forming units.
There was no visible colony forming units in the tap water or the sample taken from Vidy (the unknown). The colony forming units of E.coli counted from the Vauchère samples are presented in the table below.
Colony forming units counted Vauchere
| Counts | Vauchere 1 | Vauchere 2 | Vauchere 3 | Avg | Std Dev |
|---|---|---|---|---|---|
| Agir | 7 | 3 | 29 | 13 | 14 |
| hammerdirt | 7 | 3 | 27 | 12 | 13 |
The ECA Check Easygel material arrived on June 11th so as a control that they work as expected - 0.5ml of the same Tap Water and Vuachère (saved in the refrigerator) samples were plated. The EasyGel is much easier to distinguish the colony forming units than the methylene blue agar.
Literature
2014 Report on Leman biomass
2014 Physico-chemical Leman report
Relevant Links
Discard Studies publishes interesting articles on Water Pollution
- Pharmaceuticals Out of Bounds - conference coming up in 2016
- DIY Plastics Collector "Babylegs"
- Redefining pollution and action - article
Electronic Lab Notebooks
- A universal open-source Electronic Laboratory Notebook Bioinformatics 2013 29(13):1710-2. doi: 10.1093/bioinformatics/btt253
Documentation and Collaboration Tools
Agree on documentation and collaboration tools
- Internal communication - hackuarium slack
- Calendar? - we have a biodesign google calendar
- Protocols - [1]?
- Electronic Lab Notebook - [2]? - a template system where people can write down what they do would be nice, should it be the same as the Data entry format where you comment on unusual procedures or events?
- Data entry and sharing - Hammerdirt github, compliance with the international standards
- Hardware design - biodesign github
Water Collection Protocols
Coliform Bacteria Counting
The presence of E. coli in natural waters is a direct indication of fecal contamination of water. Coliform bacteria and other bacterial species can also be found in public waters and contribute to human pathologies. There are several methods to distinguish E. coli, coliform and other bacteria vs other microorganisms that may grow on an agar plate, of special interest 'chromogenic media' which contain substrates that form specific colors depending on enzymes produced by each species.
Easy Gel
Micrology Labs Coliscan Easygel
- Rachel asked for an offer including shipping and the 15% discount for the a coliform bacteria testing medium.
- reminder request sent again 18 April to Jonathan Roth (CEO of Micrology Labs) and Doug Wengerd (who eventually answers notes to their 'info' address)
Protocol from the Micrology Labs
- there have been some paying complications too, but their large scale order for 100 plates makes the 'per plate' cost half of another option
To note: the Micrology Lab ECA Easygel plates are somewhat expensive, but can assay up to 5ml of sampled water per plate for ready scoring and disposal. Another currently proposed plate media would not only be less discriminating, but require more equipment for the sterile setup and final cleanup, and probably only allow 0.5ml to be assayed/plate. The Micrology plates are also used in other 'citizen science' campaigns and approved by regulatory bodies...
Other Media
- Endo agar where gram negatives are favored to grow. Fermentation of lactose in the medium by coliform bacteria gives metallic green sheen to those colonies.
- MacConkey agar - also contains lactose and neutral red as a pH indicator, bile salts to inhibit gram-positive bacteria growth
- MacConkey agar with sorbitol is used to isolate E. coli O157, a pathogenic enteric bacteria
- Eosin Methylene Blue agar - Lactose fermenting bacteria turn a dark color, gram positive bacteria also inhibited
DIY protocol
Automatic Colony Counter and Plate Counting
This is not a DIY gel, but ENDO agar was used for the workshops Lifepatch conducted in Indonesia: Quantification of e. coli contamination in water
From Lifepatch, we have set this up DIY automatic colony counter.
Other Methods
- Good "travaux pratique" protocol for bacterial counting and salts concentration from Bioutils, UNIGE
- Other protocol proposing several direct and indirect methods
Biodesign has tried:
- Hack-a-Taq
- This early post has a pdf at the end summarizing techniques, including portable sensor, paper strip, etc.
Scaling
Basic calculation for cost for one time analysis:
- number of plates = number of sites x number of dilutions x number of replicates + positive control + negative control
Use glass petri-dishes for recycling - we can as Lifepatch how they set it up.
Community Bioreporter Kit for Switzerland
- Our current prototype needs to be field-ready - the final design parameters need to be discussed.
- The protocol for the bioreporter assay needs to be simplified.
Otherwise, the system for working with the bioreporters has been approved with the Swiss authorities.
Water Multi-meter
- (Art)ScienceBLR
- Student Project 2016
Press
Short History of the Collaboration
Hammerdirt came to the BIO-DESIGN for the REAL WORLD's Winter School with data on the macropollutants washing up on the local beaches in and around Montreux (CH). The data format conforms to international norms, and over two years, some patterns can be observed in the level of litter on the shores. The collaboration will be an exchange and complement to what is already on-going at Hammerdirt.
The presentation from that day can be found here: How can we work together ?
Brainstorm Results
At Open Hackuarium #88, we came up with some initial ideas.
- Data Analysis
- Image analysis and machine learning to readily tabulate the macro-pollutants collected on the beach
- synching the above data with a phone application in the internationally accepted format
- Add lake existing data (lake current, meterology, etc.)
- Water collection and Analysis
- Standardizing Collection Methods
- Water chemistry testing - turbidity, pO2, pH, conductivity, temperature
- Microbial testing - distinguish fecal contaminants and others with selective plates
- DIY free NH2
- Microparticle counting - DIY
- Heavy metal analysis DIY equipment of BIODESIGN to be more robust - or
- Make a fluorescent plate reader for higher throughput (CAUTION: still need P1 to use bioreporters here)
- antibiotic resistant bacteria (CAUTION: we cannot amplify/concentrate possible pathogens)
- Fish diseases - ask Maison de la Rivière?
- Awareness Raising - Recycling
- Recycle plastics using the extruder and use it to 3D print objects
- Biodegradation with mycelium