Difference between revisions of "HTGAA"
(HTGAA Homework week 2) |
|||
| (17 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| − | == HTGAA | + | == HTGAA == |
| − | + | From the official [http://bio.academany.org/:Link webpage]: Bio Academy is a Synthetic Biology Program directed by George Church, professor of Genetics at Harvard medical school. The Bio Academy is a part of the growing Academy of (almost) Anything, or the Academany. | |
| − | + | It's a 15 week experimental course covering the major Synthetic Biology applications today. | |
| − | Lab Task: | + | = HTGAA course 2017= |
| + | 2017 is the first year that the course is being taken by students at Hackuarium. The course is being followed by [[User:Dan | Daniel Hernandez]] and Thomas Jordan. | ||
| + | |||
| + | ==Important Links== | ||
| + | |||
| + | ===Hackuarium Specific Links=== | ||
| + | Hackuarium Official Course Page: http://bio.fabcloud.io/hackuarium_2017/ | ||
| + | |||
| + | Hackuarium Working Assignments Folder: https://drive.google.com/open?id=0B26FGn9YdUxQVmZuaTBNNWlGR0k | ||
| + | |||
| + | |||
| + | ===General Course Links=== | ||
| + | |||
| + | Course page: http://bio.fabcloud.io/2017/ This includes all the weekly lectures and homework discussions in the left-hand sidebar. At the bottom there are additional learning resources. | ||
| + | |||
| + | Students: | ||
| + | https://docs.google.com/spreadsheets/d/1WRnoyLbWejQslRvaRNfQBxJkjgmKWHNfFT9BOZqdEGM/edit#gid=0 | ||
| + | |||
| + | Curriculum: | ||
| + | https://docs.google.com/spreadsheets/d/1Xg_GtrFb8wYJVUPlrbUb6pP0dWGmkoAKU4xP6qhBKVE/edit#gid=0 | ||
| + | |||
| + | Recommended Hardware: | ||
| + | https://docs.google.com/spreadsheets/d/1iu57Ct9NiPPVxzK_PBd_JlX4dlB68pQOxFCOnThnZPs/edit#gid=0 | ||
| + | |||
| + | Inventory: | ||
| + | https://docs.google.com/spreadsheets/d/1YRWQRTBSgn1aUY-WVLG3K8riCbV-ZATtPElDXcY88gQ/edit#gid=580981494 | ||
| + | |||
| + | ==Weekly topics / Experiments== | ||
| + | |||
| + | === wk1: Principles and Practices === | ||
| + | |||
| + | Link to weekly page: http://bio.fabcloud.io/2017/principles_practices_hardware.html | ||
| + | |||
| + | |||
| + | |||
| + | === wk2: Bio Design / Plasmid digest=== | ||
| + | |||
| + | Theory class: George Church spoke about the capacity to modify biological material/organisms. | ||
| + | |||
| + | Lab Task: Design and verify restriction digest of a circular DNA plasmid by gel electrophoresis. Compare your experimental result with what you would predict from the plasmid map. | ||
| + | |||
| + | Link to weekly page: http://bio.fabcloud.io/2017/bio_design.html | ||
The protocol designed for this experiment is: | The protocol designed for this experiment is: | ||
| − | https://docs.google.com/document/d/1Rs1ElrSjuLlmIEhpxZ6856ivZJBkXE4BnWnDYquZZ0w | + | |
| + | https://docs.google.com/document/d/1Rs1ElrSjuLlmIEhpxZ6856ivZJBkXE4BnWnDYquZZ0w/edit?usp=sharing | ||
| + | |||
| + | The experiment was carried out on wednesday, 11th of of October 2017 with the help of Rachel. | ||
| + | |||
| + | The gel prepared was 20 mL of 1% agarose in 1x TAE buffer. It was prepared by dissolving 0.2g agarose in 20 mL of a 1x TAE buffer solution in the microwave. Upon full dissolution of the agarose, the sample was allowed to cool a bit, SYBR safe dye was added, and the liquid cast into an old horizontal mini gel box tray. A special 'hack' with magnets kept the comb from going too deep into the liquid, as shown. | ||
| + | [[File:Gel_Hack_HTGAA.jpg |400px|]] | ||
| + | |||
| + | The Gel result was pretty good! | ||
| + | [[File:IMG 1348.JPG |400px|]] | ||
| + | |||
| + | Size markers, are in the first lane... | ||
| + | [[File:IMG 1350.PNG |200px|]] | ||
| + | |||
| + | The plasmid ypp 3-3 (Yann Pierson's) was used for these test digests, and the further lanes on this 1% Agarose mini-gel were: | ||
| + | uncut (no enzyme, but incubated) <br> | ||
| + | KpnI digest (should linearize for 6.48Kb band, but got partial digest) <br> | ||
| + | PvuII digest (expected at least 5 bands, 93/125, 470, 762, 2062 & 2968 base pairs)<br> | ||
| + | SacI digest (should linearize also) <br> | ||
| + | KpnI+SacI (for double digest, expected 1.3 & 5.2Kb) <br> | ||
| + | |||
| + | More to come! | ||
| + | |||
| + | === wk3: next generation synthesis === | ||
| + | |||
| + | Theory task: design a gene with a detailed workflow of how to build it. | ||
| + | |||
| + | Lab task: Recode a GFP plasmid to another variant of fluorescent protein using a golden gate assembly. | ||
| + | |||
| + | Ideally this task should be started ASAP as soon as the oligos arrive for the golden gate assembly | ||
| + | |||
| + | == Headline text == | ||
Latest revision as of 10:15, 21 November 2017
HTGAA
From the official webpage: Bio Academy is a Synthetic Biology Program directed by George Church, professor of Genetics at Harvard medical school. The Bio Academy is a part of the growing Academy of (almost) Anything, or the Academany.
It's a 15 week experimental course covering the major Synthetic Biology applications today.
HTGAA course 2017
2017 is the first year that the course is being taken by students at Hackuarium. The course is being followed by Daniel Hernandez and Thomas Jordan.
Important Links
Hackuarium Specific Links
Hackuarium Official Course Page: http://bio.fabcloud.io/hackuarium_2017/
Hackuarium Working Assignments Folder: https://drive.google.com/open?id=0B26FGn9YdUxQVmZuaTBNNWlGR0k
General Course Links
Course page: http://bio.fabcloud.io/2017/ This includes all the weekly lectures and homework discussions in the left-hand sidebar. At the bottom there are additional learning resources.
Students: https://docs.google.com/spreadsheets/d/1WRnoyLbWejQslRvaRNfQBxJkjgmKWHNfFT9BOZqdEGM/edit#gid=0
Curriculum: https://docs.google.com/spreadsheets/d/1Xg_GtrFb8wYJVUPlrbUb6pP0dWGmkoAKU4xP6qhBKVE/edit#gid=0
Recommended Hardware: https://docs.google.com/spreadsheets/d/1iu57Ct9NiPPVxzK_PBd_JlX4dlB68pQOxFCOnThnZPs/edit#gid=0
Weekly topics / Experiments
wk1: Principles and Practices
Link to weekly page: http://bio.fabcloud.io/2017/principles_practices_hardware.html
wk2: Bio Design / Plasmid digest
Theory class: George Church spoke about the capacity to modify biological material/organisms.
Lab Task: Design and verify restriction digest of a circular DNA plasmid by gel electrophoresis. Compare your experimental result with what you would predict from the plasmid map.
Link to weekly page: http://bio.fabcloud.io/2017/bio_design.html
The protocol designed for this experiment is:
https://docs.google.com/document/d/1Rs1ElrSjuLlmIEhpxZ6856ivZJBkXE4BnWnDYquZZ0w/edit?usp=sharing
The experiment was carried out on wednesday, 11th of of October 2017 with the help of Rachel.
The gel prepared was 20 mL of 1% agarose in 1x TAE buffer. It was prepared by dissolving 0.2g agarose in 20 mL of a 1x TAE buffer solution in the microwave. Upon full dissolution of the agarose, the sample was allowed to cool a bit, SYBR safe dye was added, and the liquid cast into an old horizontal mini gel box tray. A special 'hack' with magnets kept the comb from going too deep into the liquid, as shown.

The Gel result was pretty good!
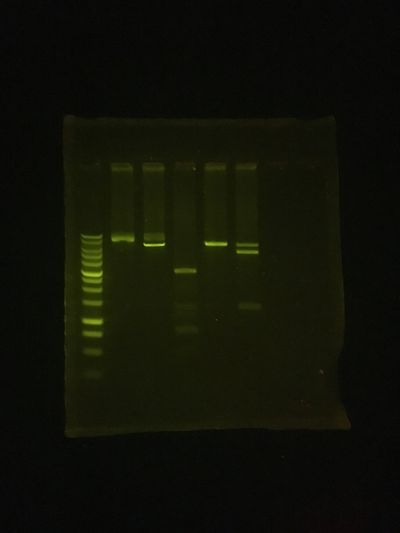
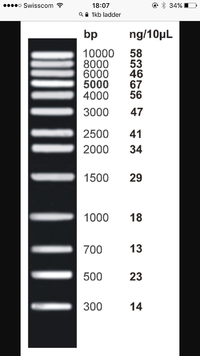
Size markers, are in the first lane...
The plasmid ypp 3-3 (Yann Pierson's) was used for these test digests, and the further lanes on this 1% Agarose mini-gel were:
uncut (no enzyme, but incubated)
KpnI digest (should linearize for 6.48Kb band, but got partial digest)
PvuII digest (expected at least 5 bands, 93/125, 470, 762, 2062 & 2968 base pairs)
SacI digest (should linearize also)
KpnI+SacI (for double digest, expected 1.3 & 5.2Kb)
More to come!
wk3: next generation synthesis
Theory task: design a gene with a detailed workflow of how to build it.
Lab task: Recode a GFP plasmid to another variant of fluorescent protein using a golden gate assembly.
Ideally this task should be started ASAP as soon as the oligos arrive for the golden gate assembly