Difference between revisions of "Bioluminescence investigations"
| (33 intermediate revisions by the same user not shown) | |||
| Line 9: | Line 9: | ||
= Who is involved?= | = Who is involved?= | ||
| − | It all started with 2 students from Nyon, who wanted to do their 'travail de maturité' with some bioluminscent | + | It all started with 2 students from Nyon, who wanted to do their 'travail de maturité' with some bioluminscent bacteria, to make a mosaic! <br> |
*Inès Nerodenkova | *Inès Nerodenkova | ||
*Luca DN | *Luca DN | ||
| Line 15: | Line 15: | ||
They ordered ''Vibrio fischeri'' from a French biotech company in [https://www.conatex.com/fr/catalog/svt/microbiologie/experiences_avec_des_micro_organismes/product-kit_bioluminescence/sku-1093487#.XMReN-szbwc a special kit]. | They ordered ''Vibrio fischeri'' from a French biotech company in [https://www.conatex.com/fr/catalog/svt/microbiologie/experiences_avec_des_micro_organismes/product-kit_bioluminescence/sku-1093487#.XMReN-szbwc a special kit]. | ||
| − | while a bit of light was visible in cultures, in comparison to controls, and determined by unbiased dark-adapted participants, it | + | while a bit of light was visible in cultures, in comparison to controls, and determined by unbiased dark-adapted participants, it was not very bright, and no good pictures were obtained of it...<br> |
Advice was requested from | Advice was requested from | ||
*Siouxsie Wiles - Australian researcher, who recommended ''Photobacterium phosphorum'', as brighter. | *Siouxsie Wiles - Australian researcher, who recommended ''Photobacterium phosphorum'', as brighter. | ||
Help was also obtained from | Help was also obtained from | ||
| − | *Hackuarium members (Rachel, Luc H) | + | *Hackuarium members (Rachel, Luc H, Gustavo, with his advice on dark adaptation, pirate-style) |
| − | *more members | + | *more members got involved (Esther) and |
| − | *new members | + | *new members (sort of) came on board (Christelle) for more fun! |
esp, as Rachel also ordered some [https://www.carolina.com/teacher-resources/Document/bioluminescent-dinoflagellates-care-sheet/tr39813.tr bioluminescent Dinoflagellates] from a company in the states. | esp, as Rachel also ordered some [https://www.carolina.com/teacher-resources/Document/bioluminescent-dinoflagellates-care-sheet/tr39813.tr bioluminescent Dinoflagellates] from a company in the states. | ||
| + | <br> | ||
| + | glow bugs in the garden during the season of corona may inspire a new project? (impossible? anyway, they may have fluor not biolum... to be confirmed) <br> | ||
| + | (see pics below - cute.)<br> | ||
| − | = | + | = Results in the BioLab in Ecublens= |
| − | + | The strain obtained by the students from Nyon last year actually isn't ''V. fischeri,'' but the recommended ''Photobacterium phosphorum'' | |
| − | *pcr | + | *pcr with 16S primers 671 and 672... <br> |
| − | + | 671: primer name UV_27F Sequence: AGR GTT YGA TYM TGG CTC AG <br> | |
| + | 672: primer name UV_907R Sequence: CCG TCA ATT CMT TTR AGT TT<br> | ||
| − | about 92% identity over the 16S rRNA region sequenced, so it seems clear that the french co also thought the ''P. phos'' was brighter, but didn't want to waste their kit boxes! :) | + | <br> |
| + | For additional documentation, 27F and 907R are in this table:<br> | ||
| + | https://www.nature.com/articles/nprot.2014.067/tables/1 <br> | ||
| + | (there are references of other classic papers to the right of this table)<br> | ||
| + | <br> | ||
| + | Here are the PCR reaction and cycling conditions we use (with Q5 pol to allow proofreading and avoid errors)<br> | ||
| + | master mix for 5 rxns (25ul total)<br> | ||
| + | 25 ul 5x Q5 buffer<br> | ||
| + | 79.375 ul H2O<br> | ||
| + | 3.125ul 10mM dNTPs<br> | ||
| + | 6.25ul Forward primer<br> | ||
| + | 6.25ul Reverse primer <br> stocks at 10uM, for each primer<br> | ||
| + | 2.5ul enz<br> | ||
| + | 24.5ul into 5 tubes... | ||
| + | add 0.5ul water in Tube 0 - negative control <br> | ||
| + | <br> | ||
| + | 4min at 94 degrees celsius<br> | ||
| + | 35 cycles:<br> | ||
| + | 20s 94<br> | ||
| + | 20s 60<br> | ||
| + | 30 s 72<br> | ||
| + | Final extension<br> | ||
| + | 2 min at 72<br> | ||
| + | |||
| + | (To note: different Taq polymerases might prefer different cycling conditions, and <br>we generally use freeze-thaw extracts of each bacterial colony, as all species do not amplify if simply added to the mix with a toothpick.<br> | ||
| + | We touch the toothpick tip to the colony to be tested, and swirl it in a tube with 20ul of water (PCR clean, of course!) and put it to freeze.<br> After thawing, 0.5ul of this solution is good input for a pcr reaction.)<br> | ||
| + | |||
| + | |||
| + | After PCR and band purification (Qiagen columns) and<br> | ||
| + | then, sequencing (light-seq from GATC, just about 5chf/result!) ,<br> | ||
| + | we obtained very clear results! <br> | ||
| + | |||
| + | blast showed about 92% identity over the 16S rRNA region sequenced vs. ''V. fischeri'', so it seems clear that the french co also thought the ''P. phos'' was brighter, but didn't want to waste their kit boxes! :) | ||
[[File:1555510582.gif]] | [[File:1555510582.gif]] | ||
[[File:Vfischerii_vs_Phosphobact.png]] | [[File:Vfischerii_vs_Phosphobact.png]] | ||
| + | |||
| + | *Christelle got some good pictures from plates, but new experiments with more salt are being run now! | ||
| + | |||
| + | [[File:Morebiolum.png | 250px]] | ||
| + | |||
| + | *This is one of the best pics of the dinoflagellates (about a half hour after their 'twilight' - they are strict in terms of circadian rhythms) so far... | ||
| + | [[File:DSC 8698.jpg | 250px]] | ||
| + | |||
| + | Will add more pics of these dino cells in this gallery: | ||
| + | [[File:TappedDinos.png|200px|thumb|left|tapped]] | ||
| + | [[File:Dinoflag plankton 100x.png|200px|thumb|left|subcellular - w/ chloroplasts happy...]] | ||
=What's Next= | =What's Next= | ||
| − | + | The challenge is to get the biolum organisms' real color(s) in images, and to make them bright... <br> extra salt is seemingly very important! <br> but maybe not [https://www.jstage.jst.go.jp/article/jgam1955/26/2/26_2_75/_pdf malic acid]... The second test for this is ongoing! <br> | |
| − | more results to come | + | Clearly, temperature is a big factor in getting colonies bright! After being in the frig, we see more!! A temperature sensitive lux enzyme (or two?) needs time in the cold to brighten up colonies.<br> |
| + | However, the brightest colonies do not always streak out well on fresh plates. Maybe the aldehydes in the light reaction are toxic, and this is part of the reason for quorum effect regulation - to make sure there are some other bugs that will not be bright, and remain able to grow!?? <br> | ||
| + | more results to come. | ||
| + | |||
| + | For instance... in 2025 we got some bioluminescent mycelia from Ove Mattmann, ''Panellus stipticus'', and did some experiments with pH with a gymnase student, Lucie D, doing her TM on bioluminescence. Here are some [https://docs.google.com/presentation/d/12R1h-IVZR3m5qv_aPtyRH_s-0a2QUC-Jy1U3kSrypcI/edit?usp=sharing first results]. Interestingly, not only did they require fresher malt extract for glowing on our plates, but their biolum comes out green on the same camera as the one I used for the dinoflagellates...[[File:Brightfield.png|thumb|to left, P. stipticus: above, normal media, below acidic. to right, F. fomentarius, a non luminescent mycelia.]] | ||
| + | [[File:Screenshotwithambientlight firstPositivepHeffect.png|alt=fades a week later|thumb|Screenshot of biolum from ''P. stipticus'' on plates with ambient light - our first Positive pH effect ]]The mycelia of the ''P. stipticus'' were sensitive to carmelized old maltose, and only brightened up in culture after we got a new batch. For another TM the pH tests were fun to initiate, with Citric Acid (1M) used for adjusting it, with buffered solutions included for the liquid based tests... | ||
| + | |||
| + | Clear effects of growth and luminescence were noted. Can see the link above for still more details... | ||
| + | |||
| + | This [https://pmc.ncbi.nlm.nih.gov/articles/PMC6294908/ article talks about the genetics of mycelia bioluminescence]. Apparently, ''P. stipticus'' has just a Luciferase and an enzyme (h3h) to make the substrate luciferin. | ||
Latest revision as of 17:08, 3 September 2025
Bioluminescence Investigations
What
We are investigating life that gives off light!!
already we have dinoflagellates (without flagella!) and Photobacterium phosphorum !
(hoping for some glowing mycelia, but wonder if it is simply phosphorescence there...)
Who is involved?
It all started with 2 students from Nyon, who wanted to do their 'travail de maturité' with some bioluminscent bacteria, to make a mosaic!
- Inès Nerodenkova
- Luca DN
They ordered Vibrio fischeri from a French biotech company in a special kit.
while a bit of light was visible in cultures, in comparison to controls, and determined by unbiased dark-adapted participants, it was not very bright, and no good pictures were obtained of it...
Advice was requested from
- Siouxsie Wiles - Australian researcher, who recommended Photobacterium phosphorum, as brighter.
Help was also obtained from
- Hackuarium members (Rachel, Luc H, Gustavo, with his advice on dark adaptation, pirate-style)
- more members got involved (Esther) and
- new members (sort of) came on board (Christelle) for more fun!
esp, as Rachel also ordered some bioluminescent Dinoflagellates from a company in the states.
glow bugs in the garden during the season of corona may inspire a new project? (impossible? anyway, they may have fluor not biolum... to be confirmed)
(see pics below - cute.)
Results in the BioLab in Ecublens
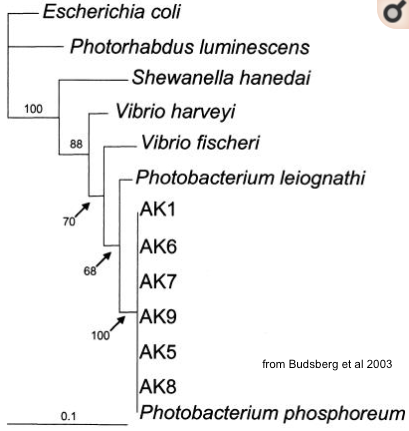
The strain obtained by the students from Nyon last year actually isn't V. fischeri, but the recommended Photobacterium phosphorum
- pcr with 16S primers 671 and 672...
671: primer name UV_27F Sequence: AGR GTT YGA TYM TGG CTC AG
672: primer name UV_907R Sequence: CCG TCA ATT CMT TTR AGT TT
For additional documentation, 27F and 907R are in this table:
https://www.nature.com/articles/nprot.2014.067/tables/1
(there are references of other classic papers to the right of this table)
Here are the PCR reaction and cycling conditions we use (with Q5 pol to allow proofreading and avoid errors)
master mix for 5 rxns (25ul total)
25 ul 5x Q5 buffer
79.375 ul H2O
3.125ul 10mM dNTPs
6.25ul Forward primer
6.25ul Reverse primer
stocks at 10uM, for each primer
2.5ul enz
24.5ul into 5 tubes...
add 0.5ul water in Tube 0 - negative control
4min at 94 degrees celsius
35 cycles:
20s 94
20s 60
30 s 72
Final extension
2 min at 72
(To note: different Taq polymerases might prefer different cycling conditions, and
we generally use freeze-thaw extracts of each bacterial colony, as all species do not amplify if simply added to the mix with a toothpick.
We touch the toothpick tip to the colony to be tested, and swirl it in a tube with 20ul of water (PCR clean, of course!) and put it to freeze.
After thawing, 0.5ul of this solution is good input for a pcr reaction.)
After PCR and band purification (Qiagen columns) and
then, sequencing (light-seq from GATC, just about 5chf/result!) ,
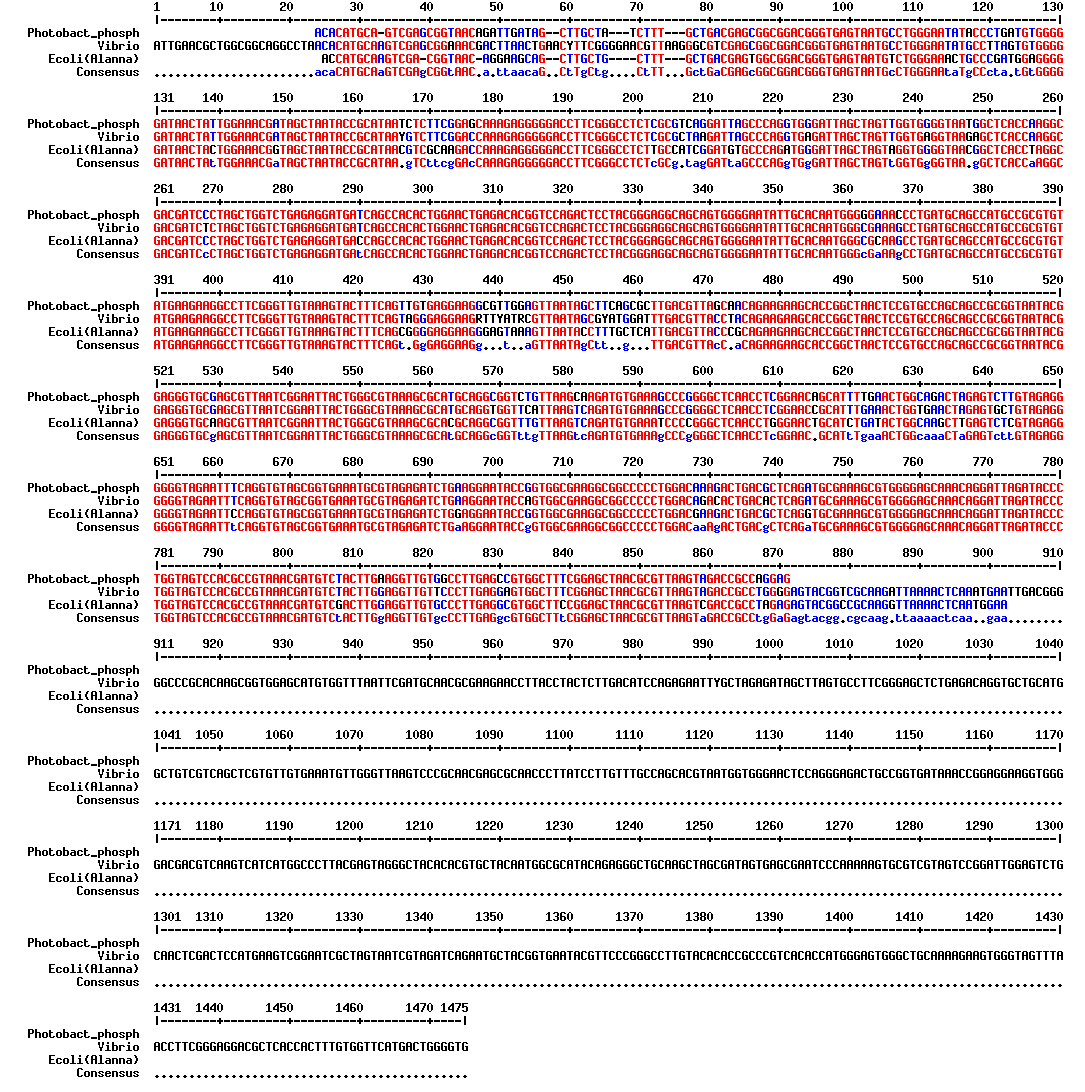
we obtained very clear results!
blast showed about 92% identity over the 16S rRNA region sequenced vs. V. fischeri, so it seems clear that the french co also thought the P. phos was brighter, but didn't want to waste their kit boxes! :)
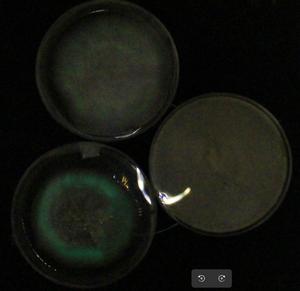
- Christelle got some good pictures from plates, but new experiments with more salt are being run now!
- This is one of the best pics of the dinoflagellates (about a half hour after their 'twilight' - they are strict in terms of circadian rhythms) so far...
Will add more pics of these dino cells in this gallery:
What's Next
The challenge is to get the biolum organisms' real color(s) in images, and to make them bright...
extra salt is seemingly very important!
but maybe not malic acid... The second test for this is ongoing!
Clearly, temperature is a big factor in getting colonies bright! After being in the frig, we see more!! A temperature sensitive lux enzyme (or two?) needs time in the cold to brighten up colonies.
However, the brightest colonies do not always streak out well on fresh plates. Maybe the aldehydes in the light reaction are toxic, and this is part of the reason for quorum effect regulation - to make sure there are some other bugs that will not be bright, and remain able to grow!??
more results to come.
For instance... in 2025 we got some bioluminescent mycelia from Ove Mattmann, Panellus stipticus, and did some experiments with pH with a gymnase student, Lucie D, doing her TM on bioluminescence. Here are some first results. Interestingly, not only did they require fresher malt extract for glowing on our plates, but their biolum comes out green on the same camera as the one I used for the dinoflagellates...
The mycelia of the P. stipticus were sensitive to carmelized old maltose, and only brightened up in culture after we got a new batch. For another TM the pH tests were fun to initiate, with Citric Acid (1M) used for adjusting it, with buffered solutions included for the liquid based tests...
Clear effects of growth and luminescence were noted. Can see the link above for still more details...
This article talks about the genetics of mycelia bioluminescence. Apparently, P. stipticus has just a Luciferase and an enzyme (h3h) to make the substrate luciferin.