Difference between revisions of "AGiR! for genomic integrity"
| Line 38: | Line 38: | ||
* Study the feasibility of further methods that could contribute to this project - i.e. an assay on either bacterial or bulk DNA | * Study the feasibility of further methods that could contribute to this project - i.e. an assay on either bacterial or bulk DNA | ||
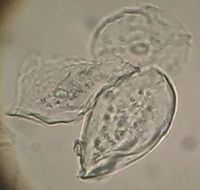
| − | Cheek cell epithelia are about 50 micrometers in diameter. [[File:Cells3.jpg|200px|thumb|right|Three cheek cells imaged on the scope at the Hackuarium. Some methylene blue stain may be visible in the most 'in focus' nucleus - left, middle cell. ]] | + | |
| + | '''Cheek cell epithelia are about 50 micrometers in diameter.''' [[File:Cells3.jpg|200px|thumb|right|Three cheek cells imaged on the scope at the Hackuarium. Some methylene blue stain may be visible in the most 'in focus' nucleus - left, middle cell. ]] | ||
== Background & Inspiration == | == Background & Inspiration == | ||
Revision as of 07:03, 18 May 2016
Genomic integrity is a new 'big picture' concept to aid public health efforts and basically includes all the molecular genetic details of cells!
So many things we commonly choose to do can impact genomic integrity, also potentially affecting future generations!
At the Hackuarium, AGiR! hopes to really get some citizen science action going - developing DIYbio methods to readily assay *your very own* cells for DNA damage, using classic (circa 1980s) techniques. If these can still be sold in 'kits,' maybe it will also be simple to make them available to all for open source efforts?
Here is the basic plan: using inner cheek cells for micronuclei and comet assays!
Watch this space for more info and visit
the AGiR! site
Introduction
Context
This is a part of a new Hackuarium project on genomic integrity.
This wiki page is under construction, and will help keep track as we develop open source simple protocols of two classic methods for quantitation of DNA damage in human cells, the micronuclei and comet assays.
People
Contact and lead:
- Rachel Aronoff
Participants:
- maybe you?!
Main Goals
Genomic integrity is a big picture concept which includes basically all the molecular genetic details of cells (from RNAs to DNAs and the epigenetic marks for proper gene expression), and thus is a topic that can be explored in several ways. For the genomic integrity project at the Hackuarium the focus will be on directly quantifying DNA damage. Comet and micronuclei assays were developed in the 1980s and publications about use of these assays on human cheek cells are already in the literature. These methods, as open source protocols, will encourage citizen science and raise awareness. Additionally, they can provide an easily "workshopable" way to show people more about their own cells, while learning how to assess DNA damage.
In other words, this experiment can be used as a gateway, to demonstrate and raise awareness for everyone on the damage possible from many common substances or activities - on your own cells, your own genetic information!
In the scope of Hackuarium activities, mammalian cell cultures are still (april 2016) a no go, but freshly isolated cheek cells from individuals can be used to do tests on DNA damaging compounds or activities. Such 'ex situ' assays might seem more attractive, especially if potentially toxic compounds are tested! (but the caveat is that it is only after the metabolism of some compounds that the DNA-damaging effects are 'revealed' - for example alcohol to DNA-damaging aldehydes...
In order to further investigate compounds that might cause DNA damage, a classic test of bacterial reversion (i.e. the Ames test) or on bulk DNA may also be nice to establish at the Hackuarium.
If enough people follow the 'open source' protocol(s) that we develop to test substances on their cheek cells, and clearly significant effects are observed, we may even be able to publish such effects.
Objectives
- Obtain ready results with cheek cells for the comet and micronuclei assays
- Put together the open source protocol, with suggestions for standard tests and controls
- Demonstrate methods and 'share the love' with other biohacker groups (La Paillasse etc.)
- Study the feasibility of further methods that could contribute to this project - i.e. an assay on either bacterial or bulk DNA
Cheek cell epithelia are about 50 micrometers in diameter.
Background & Inspiration
Here is the link to the Prezi Rachel presented (13apr16) at an Open hackuarium night! Prezi on DIYbio for Genomic integrity!
Some theory
More on comet cells and micronuclei to come soon!
But in the meantime, here are the references to the cheek cell papers mentioned in the presentation (and in the prezi):
Szeto et al (2005) Mutation Research 578:371-381
(In this paper, green tea is also shown to be protective.)
dos Santos Rocha et al (2014) Genetics and Molecular Biology, 37(4)702-707
(This paper looks at micronuclei in cheek cells (and also analyses apoptosis) from mouthwash and alcohol users.)
Inspiration / References / Similar projects
Inspiration: good pages to come
- [http://]
- [Link 2]
Setup description
General principle:
Necessary Materials
- Interested citizen scientists
- Lab equipment for the cheek cell 'comet' and micronuclei assays
- Microscope, glass slides
- Lab 'consumables' pipets and tips, beakers
- Agarose, and gel running apparatus for embedding cells, treatments (triton, protease, EDTA, buffer solutions, etc.) and nucleoid migration (for the comet assay)
- Mercury Lamp Setup and filters for fluorescent dye detection.
- Could also try to design a transilluminator suitable for cellular resolution (UV/Fluo as usual for comets) when imaging (to look for open source models)
- Other reagents and histochemical stains (methylene blue)
- Hydrogen peroxide for positive control cells (oxidative damage)
- Antioxidant molecules as possible 'protective' intervention
- A toothbrush for gentle collection of the cheek cells
- More details to come (What, size | quantity | where to get it | price / unit)
Parameters for the Open Source Protocol
- To get baseline conditions first and compare with the standard lab technique
then To vary | Values
- p1 |
- p2 |
- Constants (Key elements of set-up)
- c1 |
- c2
- To measure/ observe
- m1
- m2
Protocol (Plan)
Startup
Phase 1
do what | frequency | duration
Lab Notebook & News (Real)
Protocols and measurements to be linked from here: [link to other doc]
Run / prototype #2
What's new compared to #1: