Difference between revisions of "Pushing the P1"
| (88 intermediate revisions by 6 users not shown) | |||
| Line 1: | Line 1: | ||
| − | = | + | =A P1 Lab for Open and Accessible DIT-Research = |
| − | Having a laboratory and instruments is one thing, possessing the tools to do actual biology research is another thing. For | + | Having a laboratory and instruments is one thing, possessing the tools to do actual biology research is another thing. For over three years, the Hackuarium community has built a space where people can work on projects related to biology among other fields. The ambition of the international DIY biology scene is to bring to the people the tools to perform and work on the whole range of biological applications, as qualitatively and cheaper than what the industrial or academic institutions do. To achieve such level, the Hackuarium lab has finally been renovated, to include a space that is upgraded to a higher level of competency, for experimentation at the P1 biosafety level where genetic manipulation opens the door to a vast range of research topic and engineering opportunities.<br> |
| − | + | We plan P1 work to start out in bacteria. Depending on future demands, we can imagine adding other activities.<br> | |
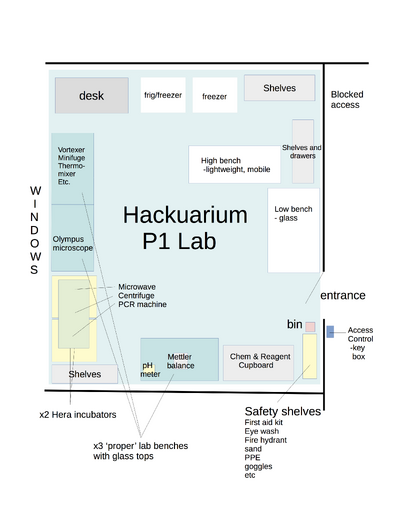
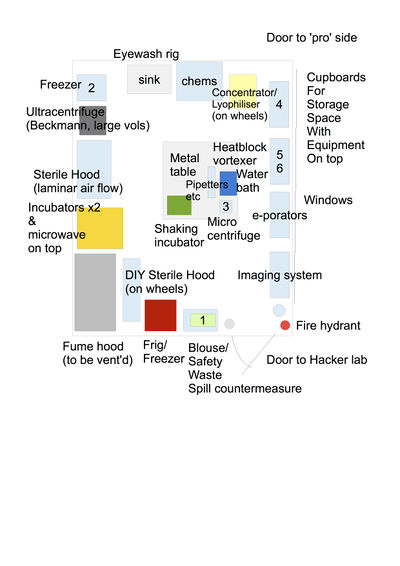
| + | [[File:P1lab2023.png|400px|thumb|center|The current P1 Lab in the Coop Space in Ecublens.]]<br> [[File:P1labplan version0.2 270917.jpg|400px|thumb|center|THE P1 LAB CONFIGURATION back in Renens (ca. 2018)]]<br> | ||
| + | <br> | ||
| + | ==Previous situation== | ||
| + | The authorities were initially notified that our P1 activities would officially begin on 1november2017.<br> | ||
| + | Acknowledgement of this info has been [https://drive.google.com/open?id=0B1jaPHOkH_42ODVtY2NNSVF0TUk received from bafu], but we still need to organise a few more things in the lab, finalise the map, and find a deputy biosecurity officer to help Rachel. (initially this was Olivier, and later Javier took on this role)<br> | ||
| + | Additionally, getting this documentation also in French could be quite important for everyone to benefit. <br> | ||
| + | (thanks in advance for some help!) <br> | ||
| + | |||
| + | In the current context (october 2017) of the P1 lab on the 2nd floor, we started again with the HTGAA course, for which some [http://wiki.hackuarium.ch/w/HTGAA description of activities] by Daniel and Thomas is currently being put into the wiki. <br> | ||
| + | Rachel, as acting biosecurity officer, has already gotten Hackuarium into the FOEN database, and has notified them that the official start of P1 activities (in particular, selection of clones for the HTGAA to begin) will be 1 November 2017. | ||
| + | Additionally, 6 people (including Dan and Thomas) have currently taken the intial P1 user intro to biosafety course, discussed further below. | ||
| + | |||
| + | '''Old Renens News (update, 1nov17)''' | ||
| + | Our first bacterial strains are now growing on plates in the incubator! <br> | ||
| + | Looking forward to the seeing bioluminescence from one of these strains, and fluorescence (esp red) from the other! <br> | ||
| + | |||
| + | We have more recent pages around our P1 activities... but this next should still be correct. | ||
| + | [https://docs.google.com/spreadsheets/d/1xPDdciRnwBRrdXu_wT6FX3BfO8Sp1qZ6EAoU31n5PQc/edit?usp=sharing Here] is the current list of strains in the lab.<br> | ||
| + | [https://docs.google.com/spreadsheets/d/1EwJ2i03dDeRm01-yQTxCNMZSTpsEWj0K418y1FhTjLo/edit#gid=0 Here] the current list of P1 lab users. <br> | ||
| + | If you have a project idea for the lab, make sure to get trained! <br> | ||
| + | Talk to the biosafety officer, Rachel Aronoff, and deputy biosafety officer, Olivier Emery, about your plans in advance! <br> | ||
| + | Documentation on the wiki is highly encouraged. <br> | ||
| + | |||
| + | ==Abbreviations == | ||
| + | Biological safety level 1 lab (P1), biosafety officer (BSO), standard operating procedure (SOP)... | ||
| + | |||
| + | ==Microbes== | ||
| + | We now have the right to work with organisms classified as BSL1 according to Swiss Law. | ||
| + | |||
| + | The first step is to check if the microbe you want to work is BSL1, go to https://www.bafu.admin.ch/bafu/fr/home/themes/biotechnologie/publications-etudes/publications/classification-des-organismes.html | ||
| + | to find the official Swiss classification. | ||
| + | |||
| + | The second step is to borrow the microbe from someone that already used it before. All BSL1 manipulations are announced to the government that makes the database freely accessible here: http://www.ecogen.ch/ecogen/Forms/Register/RegisterSearch.aspx go and find a researcher. Talking with someone that has experience on that specific microbe will be useful also to get some tips and tricks on growing conditions, etc.. | ||
| + | |||
| + | If nobody worked with the microbe before. It is possible to purchase a culture from the Culture Collection of Switzerland (CH): https://www.ccos.ch/ | ||
| + | |||
| + | Additional repositories: | ||
| + | *Algae and protozoa (UK): https://www.ccap.ac.uk/ | ||
| + | *Eukariotic cells and microbes (US):ATCC https://www.lgcstandards-atcc.org/?geo_country=ch Note: the ATCC BSL classification is American. It may differ from the Swiss one. Check the Swiss one before! | ||
| + | *Czech Collection of Microorganisms (CCM) http://www.sci.muni.cz/ccm/index.html | ||
| + | |||
| + | ==Primers== | ||
| + | * [http://www.microsynth.ch/ Microsynth] : ~ 10.-/30 bp primer | ||
| + | * Eurofins | ||
| + | * IDT | ||
| + | |||
| + | ==Sequencing== | ||
| + | *[http://www.gatc-biotech.com/fr/lp/sequencage-nextgen.html?gclid=CKzRrtTbm80CFdiZGwodBycOaA GATC] | ||
| + | *Eurofins Genomics | ||
| + | |||
| + | ==Genes== | ||
| + | A strain carrying the full luciferase operon, and a strain carrying a GFP plasmid induced by arsenic, and a constitutive mCherry plasmid, are the first being grown in the P1 lab (1nov17). <br> | ||
| + | A synthetic biology plasmid made by a golden gate multipart ligation is the next strain expected. (to make a gfp plasmid become blue...) | ||
| + | |||
| − | = | + | ==Following Norms== |
| − | + | <br><br> | |
| − | [ | + | ===Define the planned experiments (Daniel for HTGAA Academy, Rachel bio-luminescence)=== |
| − | The | + | ===Describe training program "Intro to Biosafety"=== |
| + | A [https://drive.google.com/open?id=0B1jaPHOkH_42dVhjOXBBN0xsUms Hackuarium Drive folder] with various handouts for the 1.5h intro course, and further biosafety information contains much of the information about the P1 course. Additionally, a [https://prezi.com/2da7to3emhqu/basic-lab-safety/ prezi on basic laboratory safety] is recommended viewing before the course. <br> | ||
| + | P1 training includes theoretical and practical aspects of basic safety and aseptic techniques. <br> | ||
| + | Lists of P1 users and engineered or synthetic strains will be kept in the same shared 'Drive' folder. <br> | ||
| + | The first training is being held 6 October 2017, and notification of activities to Swiss authorities will follow as final lab details are sorted (ventilation of fume hood, water to sink).<br> | ||
| + | In addition to fulfilling this training, project details must be documented in the wiki, and users must also sign a waiver and acknowledgement of risks and liability before starting any experimentation in the P1 lab. <br> | ||
| + | (folder of signed waiver scans to be put in the shared folder also) <br> | ||
| + | |||
| + | =Generalized procedure= | ||
| + | These procedure will be implemented in the lab as first benchmark in order to have a working basis. <br> | ||
| + | The "project request" procedure will be a way to ensure complete transparency and applicability. It will have to be filled before any project can start by any member willing to perform GMO activities. <br> | ||
| + | |||
| + | They are optimized for cost & material efficiency. | ||
| + | ==Maximal Culture Size== | ||
| + | Technically the space does not allow us yet to use flasks over a volume of 500ml (incubator volume). A culture grows well at a fifth of the total volume (oxigenation requisit) which theoretically makes it 100ml culture. Safety wise, using a rather small enclosed space, spills have to stay managable. To avoid bacterial spread outside of the enclosure a maximal volume of populated medium of 200ml per flask is allowed. A maximal volume of 1L in total, populated or sterile medium, is allowed. | ||
| + | |||
| + | ==Spill management== | ||
| + | Droping a vial containing liquid is common. In a lab if not prpoerly handled a spill can have more dramatic effects than droping your milk at home. Here we depict the SOP for the 4 types of spills that happen with their respective anti-spill kits; acidic, basic, contaminated and neutral spills. | ||
| + | |||
| + | *1. Alert your collaborators | ||
| + | *2. Unplug and turn-off all proximate electric equipment. | ||
| + | *3. Follow the procedure describing your situaion: | ||
| + | :: | ||
| + | '''Acidic spill'''<br> | ||
| + | [https://wiki.hackuarium.ch/w/Mercury_spill Mercury Spill] | ||
| + | <br> | ||
| + | |||
| + | ==Project Request== | ||
| + | Develop a document/wiki page as template where people will have to explain the GMO they want to produce. <br> | ||
| + | As an idea it could be a form or a wiki page that '''anybody''' could see structured as the following | ||
| + | *Goal: | ||
| + | :: - What is the purpose of the GMO / protein you want to create OR: What question should it help you answer | ||
| + | ::: ex: - Sense endocrine disruptors / Produce methane / Produce dyes | ||
| + | *How does it help reach your goal | ||
| + | ::: ex: - This enzyme is known to biochemically produce red dye (add source) | ||
| + | ::: ex: - The combination of protein A,B&C could produce a sensor... | ||
| + | *Plasmid | ||
| + | :: - Tell which plasmid you will use (for archive purpose) | ||
| + | :: - Give the code of the cassette you will use | ||
| + | :: - Give the peptide translation of such cassette | ||
| + | *Template | ||
| + | :: - If you want to use several building blocks for you proteins, give details on the templates | ||
| + | *Primers | ||
| + | :: - Give the primers you are going to use to realise your clones | ||
| + | |||
| + | == Storage and Nomenclature == | ||
| + | A recurrent behavior, often observed in institutionalized spaces, where the followup on ones item naming and storage procedure is inexistant, is the massive loss of time to find a special item belonging to a past member. Here we describe a strict naming procedure for items that is, we think, unambiguous and still logical. | ||
| + | |||
| + | |||
| + | =Original Timeline (circa 2016)= | ||
| + | |||
| + | The original timeline had the following steps: | ||
| + | |||
| + | Mise en place équipe travail + évaluation des points critiques (cout, autorisations) 1 mois | ||
| + | |||
| + | Mise en place de laboratoire + obtention du materiel 1 mois | ||
| + | |||
| + | discussion avec les autoriteés et obtention des autorisations 2 semaines | ||
=Critical Points= | =Critical Points= | ||
| + | <br> | ||
'''What we need to sort out before we start:''' | '''What we need to sort out before we start:''' | ||
| + | |||
| + | This scientifically enriching, yet delicate step, is now in motion but in order to start practicing several items involving legal, technical and community topics have to be settled. Indeed, our laboratory practices and transparency as a citizen lab have to be absolutely irreproachable. The challenges we faced are described in this document.<br><br> | ||
| + | Regarding Manuela Ocaña (Collaboratrice scientifique, PhD, Département fédéral de l'intérieur DFI, Office fédéral de la santé publique OFSP):" Les instituts, les entreprises et les organisations qui utilisent des organismes génétiquement modifiés, pathogènes ou exotiques en milieu confiné sont ainsi tenus de notifier leur activité (classes 1 et 2) ou de demander une autorisation (classes 3 et 4)." Nous allons pas depasser la classe 2, du coup nous avons besoin de faire une notification (voir dessous)<br> | ||
'''Announce the first activities''' <br> | '''Announce the first activities''' <br> | ||
| − | : | + | It was done with this form [https://www.bafu.admin.ch/bafu/fr/home/themes/biotechnologie/info-specialistes/activites-en-milieu-confine/notifications-et-demandes-d-autorisation.html here] |
'''Will the enclosure be satisfactory?''' | '''Will the enclosure be satisfactory?''' | ||
| − | + | We now have a perfectly closed room with 2 doors and keys. | |
'''Do we need a bio safety officer? Does this person need a specific training?''' | '''Do we need a bio safety officer? Does this person need a specific training?''' | ||
| − | + | Yes, Rachel has set up a P2 lab before. She will nevertheless do another round of training and she designed an initial training program for users. <br> | |
'''Risk assessment''' | '''Risk assessment''' | ||
| − | ::*The biological risk assessment will be made through the form each member | + | ::*The biological risk assessment will be made through the form each member has to fill when he creates a new genetically modified strain to ensure no pathological or harmful gene is inserted in the micro-organism. |
::*Chemically, any inflammable liquid will be stored in the solvent cabinet. The acids and bases will be separated in two "geographically" remote cabinets. The bottles will be stored close to ground level in a resistant container able to contain the full volume of the bottles it contains to avoid spillage. | ::*Chemically, any inflammable liquid will be stored in the solvent cabinet. The acids and bases will be separated in two "geographically" remote cabinets. The bottles will be stored close to ground level in a resistant container able to contain the full volume of the bottles it contains to avoid spillage. | ||
| − | '''Waste | + | '''Waste management''' |
:: -> OK (see below) | :: -> OK (see below) | ||
| Line 73: | Line 188: | ||
https://docs.google.com/spreadsheets/d/1tBvxEk4UVtvzfeW_-yR9yNbrLmC1V9IDjAPit37TzAg/edit#gid=1606970814 | https://docs.google.com/spreadsheets/d/1tBvxEk4UVtvzfeW_-yR9yNbrLmC1V9IDjAPit37TzAg/edit#gid=1606970814 | ||
| − | ==Waste | + | Material to be used for HTGAA 2017 is here: |
| + | https://docs.google.com/spreadsheets/d/1Xg_GtrFb8wYJVUPlrbUb6pP0dWGmkoAKU4xP6qhBKVE/edit#gid=0 | ||
| + | |||
| + | ==Waste elimination== | ||
Here we describe who can and accepts to eliminate our wastes at what cost and try to estimate the frequency of elimination. | Here we describe who can and accepts to eliminate our wastes at what cost and try to estimate the frequency of elimination. | ||
We base the frequency on a per project base. How many liters of cultures are produced per production? How many biowaste bag can be filled in a week? etc... | We base the frequency on a per project base. How many liters of cultures are produced per production? How many biowaste bag can be filled in a week? etc... | ||
| Line 89: | Line 207: | ||
==Calculated base costs== | ==Calculated base costs== | ||
| − | *Here we | + | *Here we estimated the cost of starting the whole setup, based on the output of a reference protein laboratory's costs. |
| − | *We | + | *We also took a standardized method which is documented lower (protocol) based only on biology with ''E.coli''. |
| − | *The costs are detailed on the doc below and | + | *The costs are detailed on the doc below and calculated on the academic standards (ca. 2015 calculations). |
| − | *The goal then is to find hacks and shortcuts to largely cut the costs and make it affordable. | + | *The goal then is to find hacks and shortcuts to largely cut the costs and make it affordable. |
| + | *This exercise still needs to be redone for the 2017 P1 context. | ||
https://docs.google.com/spreadsheets/d/1tBvxEk4UVtvzfeW_-yR9yNbrLmC1V9IDjAPit37TzAg/edit#gid=0 | https://docs.google.com/spreadsheets/d/1tBvxEk4UVtvzfeW_-yR9yNbrLmC1V9IDjAPit37TzAg/edit#gid=0 | ||
| − | == | + | ==Software and Databases== |
Here we discuss the gene editing softwares to design plasmids, primers and gene building blocks. A total transparency has to be implemented in order for people to follow our activities. What kind of database will we use to store the ordered primers sequences? Same for the plasmids? | Here we discuss the gene editing softwares to design plasmids, primers and gene building blocks. A total transparency has to be implemented in order for people to follow our activities. What kind of database will we use to store the ordered primers sequences? Same for the plasmids? | ||
| − | === | + | ===Software=== |
*[http://www.geneious.com Geneious] powerful but the file format is not opensource or genebank | *[http://www.geneious.com Geneious] powerful but the file format is not opensource or genebank | ||
*[http://www.snapgene.com/products/snapgene_viewer/ Snapgene] never tried, anyone ? | *[http://www.snapgene.com/products/snapgene_viewer/ Snapgene] never tried, anyone ? | ||
| + | *One of the more recent (oct2017) on-line tools for molbio work is called [https://benchling.com/academic Benchling], which can also be used for an on-line lab notebook. This tool is currently being used in the context of the HTGAA course. | ||
===Database=== | ===Database=== | ||
| Line 106: | Line 226: | ||
==Providers== | ==Providers== | ||
Here we discuss the possible service providers, their costs and if they are ready to deal with us. We canot create plasmids without primers or genes and therefore need providers. We cannot confirm the authenticity of our work without a verified plasmid sequence and therefore need sequencing services. | Here we discuss the possible service providers, their costs and if they are ready to deal with us. We canot create plasmids without primers or genes and therefore need providers. We cannot confirm the authenticity of our work without a verified plasmid sequence and therefore need sequencing services. | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
==Legal== | ==Legal== | ||
| − | + | Backup of the first exploration document : http://wiki.hackuarium.ch/w/Talk:Pushing_the_P1#Old_Hackpad_Page (Most of the necessary infos can be found here)<br> | |
| − | As said previously our laboratory practices have to be irreproachable. We carry the "hacker" label and therefore, for the public opinion and the built of trust | + | As said previously our laboratory practices have to be irreproachable. We carry the "hacker" label and therefore, for the public opinion and the built of trust. We need to study in depth several aspects of the legislation governing genetic manipulation. |
===WHO=== | ===WHO=== | ||
The very good lab safety manual right [http://www.who.int/csr/resources/publications/biosafety/WHO_CDS_CSR_LYO_2004_11/en/ here]<br> | The very good lab safety manual right [http://www.who.int/csr/resources/publications/biosafety/WHO_CDS_CSR_LYO_2004_11/en/ here]<br> | ||
| Line 145: | Line 259: | ||
===BioSafety Officier (BSO)=== | ===BioSafety Officier (BSO)=== | ||
| + | a few members from the community did the course below...<br> | ||
Courses : http://www.bafu.admin.ch/biotechnologie/01744/02964/index.html?lang=f | Courses : http://www.bafu.admin.ch/biotechnologie/01744/02964/index.html?lang=f | ||
Infos: | Infos: | ||
| Line 153: | Line 268: | ||
Coûts: 550 francs par personne<br> | Coûts: 550 francs par personne<br> | ||
Langue: anglais<br> | Langue: anglais<br> | ||
| + | update 2017, RA will be BSO for new P1, train new users, and sign up for a refresher course soon. <br> | ||
| + | She has made some edits on the P1 lab [http://wiki.hackuarium.ch/w/P1_SOP SOP aka Standard Operating Procedure] (still a work in progress) and did the notification of activities! <br> | ||
===Annonce confédération=== | ===Annonce confédération=== | ||
Through the [http://www.bafu.admin.ch/biotechnologie/01744/03225/index.html?lang=fr BAFU portail] | Through the [http://www.bafu.admin.ch/biotechnologie/01744/03225/index.html?lang=fr BAFU portail] | ||
| − | === | + | ===Train the users=== |
| − | + | ===Put it a plan that authorizes the new people to come in=== | |
| + | ===Write the notification to the Biosafety Authorities=== | ||
| − | = | + | === Users === |
| − | + | Oppositely to the Hackuarium's "P0" lab where users can come and experiment in the greatest of liberties the P1, as previously stated, will be restricted to card holders having followed an initiatory course. Each user will be registered in a database and will chose a three letter OR digit code to identify items made by himself. Example: | |
| − | + | * John Doe : joh | |
| − | + | * Martin Luther King : mlk | |
| − | + | * George Junior : gj2 | |
| − | + | This three letter acronym is practical as it is short enough to be stuck on small tubes with a descriptor i.e: joh_p7 (John Doe Plasmid 7) | |
| − | == | + | === Descriptors === |
| − | + | In the process of producing for example a protein from scratch (from synthetic DNA) only a few items have to be stored and conserved for further use. These items often in the forms of solutions rarely exceed volumes of 200ul. Therefore the whole process of protein engineering and plasmid creation can hold in 1.5ml tubes stored in 96well cardboard freezer boxes. We list what we extimate should be conserved for further uses by other collaborators and give them a single letter code. | |
| − | + | * project/task/job : j | |
| − | * | + | * DNA segments : d |
| − | : | + | * primers/amorces : a |
| − | :: | + | * plasmids : p |
| − | * | + | * proteins/constructs : c |
| − | + | * bacteria/strain : b | |
| − | + | These six items are the valuable items to be stored. The project (j) item might seem counter intuitive but makes sense in the whole nomenclature system. Let's give some examples with the hypothetical John Doe project 41 (i.e joh_j41). | |
| − | * | + | *He might do a plasmid (joh_j41_p1) that sadly he failed so he produced four more for sequencing (joh_j41_p2, joh_j41_p3, joh_j41_p4, joh_j41_p5). |
| − | + | *The plasmid was a construction of a vector (joh_j41_dV) and two inserts (joh_j41_dI1, joh_j41_dI2) | |
| − | + | *The latter were made with six primers (joh_j41_a1, joh_j41_a2, joh_j41_a3, joh_j41_a4, joh_j41_a5, joh_j41_a6) | |
| − | + | *He finally made it and expressed his protein (joh_j41_c1) (Usually you only make a protein from a plasmid so if you re-express, as a 3rd batch, it later on: joh_j41_c3) | |
| − | * | + | The nomenclature might appear cumbersome at some point but it should make more sense once the final ''project draft'' is presented. |
| − | |||
| − | |||
| − | |||
| + | === Storage === | ||
| + | When you're in the working process, the following behavior is observed: You open the freezer, look for the box containing the items of interest (generally you have a primer box, a plasmid box, etc.), open the box and in those 96 tubes you want to find them by looking at the top of them. The "morphology" of a 1.5ml tube gives you only two options: You can write on the top only limited information. Forget about markers they will fade away unless you add transparent tape on top (which is boring). Or you can add stickers on the side of the tube that can contain more details on the content. So the following storage procedure will be observed:<br> | ||
| + | *Boxes will only contain a single type of items such as primers, plasmids or proteins. <br> | ||
| + | *Each item of whatever sort will have a unique 4 letter/digit hex code preceded by the type of item. That's roughly 65500 possibilities. This code is preceded by the adequate descriptor. Example: joh_j41_p1 (the plasmid N°1 of John's project 41) was given '''by the system''' the code p019A this is 5 letters which perfectly fit on a 1.5ml tube top if printed on a sticker. Again this looks cumbersome know but will make more sense later since the system gives and stores all the info for you. | ||
| + | === Job page === | ||
| + | We presented previously the request form. This form generates for you a wiki project page or whatever online lab-book that we still have to find. The final idea is that on the server the access to these code and data is easily accessible from a terminal in the P1 lab. | ||
| + | Here is an hypothetical [[hypo_proj | '''project job page''']] and how it should look like in the end '''after the form has been filled'''.<br> | ||
| + | When somebody works and experimentally gets the product design then he can directly fill the page or the online lab-book that should transfer the data. | ||
| + | A final page, with experimental data filled in should look [[hypo_proj_pract | '''like this''']] <br> | ||
| + | If a page is filled with such details, reproducibility and experimental follow-up is much easier. The important point is having a strong automation of the main points so as to avoid time loss of creating manually those files (took me more than 30 min making a complete one by hand, it should not take more than 10 and should be done while doing the experiment). <br> | ||
[http://wiki.hackuarium.ch/w/Form:GMO_Project » Project request form with above points is now here]. | [http://wiki.hackuarium.ch/w/Form:GMO_Project » Project request form with above points is now here]. | ||
| Line 196: | Line 321: | ||
*Good old openTaq | *Good old openTaq | ||
*DNA fragment purification by gel extraction. (cheap, easy, no need of DPNI enzymes) | *DNA fragment purification by gel extraction. (cheap, easy, no need of DPNI enzymes) | ||
| − | *Ligation through modified Gibson (lysed bacterial supernatant + enzymatic complementation (Exonuclease mostly), To investigate further see the [[Media:SLiCE.PDF | SLiCE]] method) | + | *Ligation through modified Gibson (lysed bacterial supernatant + enzymatic complementation (Exonuclease mostly), To investigate further see the [[Media:SLiCE.PDF| SLiCE]] method) |
*Transformation in chemo competent cells by heat shock (way cheaper than electro-competent cells) | *Transformation in chemo competent cells by heat shock (way cheaper than electro-competent cells) | ||
*Usual LB-agar plating with ampicilin selection | *Usual LB-agar plating with ampicilin selection | ||
| Line 240: | Line 365: | ||
Yeast specialists please provide a protocol applicable here, keep in mind cost & material optimization. | Yeast specialists please provide a protocol applicable here, keep in mind cost & material optimization. | ||
| − | = | + | =Old Plan for P1 from 2016 - OP Greenhouse Effect= |
| − | + | Building the P1 in a green house... I can already smell ideas fresh as spring flourishing in there... | |
| − | + | ||
| − | + | ==Idea== | |
| − | * | + | This clever and original idea was brought to us by our master of "let's just do it it's gonna work anyways" Gustavo, congratulation to him!<br> |
| − | * | + | Therefore we can buy a greenhouse for a reasonable price and set it up as a P1. The size will be indeed smaller but will be enough to accomodate the transformation, growth, cell lysis and waste storage until elimination.<br> |
| − | + | Several greenhouses models are presented with a rationale on which one to pick. <br> | |
| − | + | The end of the document is supplemented with tasks and points to investigate <br> | |
| − | * | + | |
| − | * | + | ==Greenhouses== |
| − | * | + | We indeed aim for the best size/price ratio.<br> |
| − | * | + | *Serre Delia, Hornbach, 11m2, 1399.- [https://www.hornbach.ch/shop/Serre-Delia-6-Allin-256x434-cm-aluminium/5624313/article.html link] |
| − | * | + | :: Initial idea |
| − | * | + | :: (-) Expensive |
| − | * | + | :: (+) Higher at the wall/roof junction (1m40) than the others. Still 1m40 is not very high. <br> |
| + | *Serre Vida XL, 12.25m2, VidaXL, 399.- [https://fr.vidaxl.ch/p/40193/serre-1225-m-alu-polycarbonate link] | ||
| + | :: (+)Free delivery | ||
| + | :: (+)Cheap | ||
| + | :: (-)Low at the wall/roof junction (95cm). BUT! For the price we can get 2 and add the two walls on top of the other giving 1.9m!. <br> | ||
| + | |||
| + | ==Building Plan==<br> | ||
| + | (the updated Draft is above, a complete version of this with measure a nice pinky shinny colors will follow the next days)<br> | ||
| + | |||
| + | ==Tasks== | ||
| + | ===General=== | ||
| + | *1. Members validate the idea or propose other ideas until Wed. (DONE) | ||
| + | *2. Ordering and building all together on the 2nd of Oct. (MATERIAL RECEIVED) | ||
| + | *3. Building zone cleaned (DONE) | ||
| + | |||
| + | ===Material & Orders=== | ||
| + | *1. Rearrange lab space (DONE) | ||
| + | *2. Think about a list of vital chems and consumables that are needed and could be stored in the P1 for easier access. | ||
| + | :: look at: https://docs.google.com/spreadsheets/d/1tBvxEk4UVtvzfeW_-yR9yNbrLmC1V9IDjAPit37TzAg/edit#gid=0 and select the vital items. | ||
| + | *3. Get labcoats of various sizes, get hooks for those | ||
| + | *4. Get a set of safety goggles | ||
| + | *5. Find fridge + -20 freezer | ||
| + | *6. Order desinfectant dispenser | ||
| + | *7. Get 3x6 multiplugs | ||
| + | *8. Card access system ( can use yann's raspi3) | ||
| + | *9. Get sturdy, closable bin for solid biowaste | ||
| + | *10. Bin for classic wastes | ||
| + | *11. Sam, get a hell stock of printable stickers (we're gonna label the shit out of everything) | ||
| + | *12. Organize one cupboard with: | ||
| + | :: pcr purif kits | ||
| + | :: miniprep kits | ||
| + | :: gel purif kits | ||
| + | :: 50, 12 & 1.5 ml plastic tubes | ||
| + | :: 1, 200 & 1000ul tips | ||
| + | :: his tag purification buffers | ||
| + | *13. On the sterile hood, find a plexiglass that we can fix on and that will cover 2/3 of the front pannel (i.e. Will make a separation betwwen the user and the working environement | ||
| + | *13. Order a pipetor on SMIPLES / ricardo for serological pipettes | ||
| + | :: https://www.fr.ricardo.ch/acheter/bureau-et-commerces/autres-metiers/laboratoires/pipette-17/v/an820658344/ | ||
| + | *14. We have two benches with the glass tops: one is not held with screws, fix it, buy long enough screws (obi) | ||
| + | *15. Clean a set of micropipettes | ||
| + | |||
| + | ===Safety=== | ||
| + | *1. Announce our first activity on the confederation website (Yann) | ||
| + | *2. Design the safety standard operating procedures (SOP) such as: | ||
| + | :: MSDS for common chemicals | ||
| + | :: What to do if a spillage of contaminated liquid occurs (how to clean it). | ||
| + | :: What to do if you get injured working with bacteria. | ||
| + | :: How to eliminate wastes, liquid and solid. | ||
| + | :: Standard lab behavior (blouse, glasses, gloves) | ||
| + | :: Where to store: acids, bases, solvents -> Rules for using each of them (how bases and acids have to be separated separately) | ||
| + | :: A very good example : http://ehs.berkeley.edu/standard-operating-procedures | ||
| + | *3. Print the following decals | ||
| + | :: Biohazard 2x | ||
| + | :: Authorized personnel only | ||
| + | :: "Wash your hands when done" | ||
| + | :: "Max X users inside" (X to be defined) | ||
| + | *4. Find an easy an accessible way to put blouses and glasses (gloves can be stored and boxes left above the bench) | ||
| + | *5. Make instrument procedures (print and put in a folder) | ||
| + | *6. Questions? There is a service in the DIYBio community for [http://ask.diybio.org/ Ask a Biosafety Professional] | ||
| + | |||
| + | ===Ideas & Concerns=== | ||
| + | *As the name says it's a greenhouse, how do we make sure it doesn't become super warm ? Can we make a cheap air cycling system ? | ||
| + | |||
| + | ==Lab Setup== | ||
| + | An older idea: Current setup higher up on this page | ||
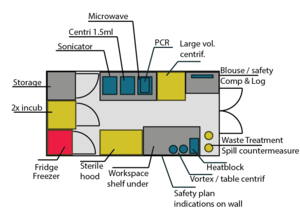
| + | [[File:GH P1.PNG|300px|thumb|center|the essential in 12m2]] | ||
| + | |||
| + | ==Next Steps - in progress as above== | ||
Latest revision as of 05:23, 3 October 2023
A P1 Lab for Open and Accessible DIT-Research
Having a laboratory and instruments is one thing, possessing the tools to do actual biology research is another thing. For over three years, the Hackuarium community has built a space where people can work on projects related to biology among other fields. The ambition of the international DIY biology scene is to bring to the people the tools to perform and work on the whole range of biological applications, as qualitatively and cheaper than what the industrial or academic institutions do. To achieve such level, the Hackuarium lab has finally been renovated, to include a space that is upgraded to a higher level of competency, for experimentation at the P1 biosafety level where genetic manipulation opens the door to a vast range of research topic and engineering opportunities.
We plan P1 work to start out in bacteria. Depending on future demands, we can imagine adding other activities.
Previous situation
The authorities were initially notified that our P1 activities would officially begin on 1november2017.
Acknowledgement of this info has been received from bafu, but we still need to organise a few more things in the lab, finalise the map, and find a deputy biosecurity officer to help Rachel. (initially this was Olivier, and later Javier took on this role)
Additionally, getting this documentation also in French could be quite important for everyone to benefit.
(thanks in advance for some help!)
In the current context (october 2017) of the P1 lab on the 2nd floor, we started again with the HTGAA course, for which some description of activities by Daniel and Thomas is currently being put into the wiki.
Rachel, as acting biosecurity officer, has already gotten Hackuarium into the FOEN database, and has notified them that the official start of P1 activities (in particular, selection of clones for the HTGAA to begin) will be 1 November 2017.
Additionally, 6 people (including Dan and Thomas) have currently taken the intial P1 user intro to biosafety course, discussed further below.
Old Renens News (update, 1nov17)
Our first bacterial strains are now growing on plates in the incubator!
Looking forward to the seeing bioluminescence from one of these strains, and fluorescence (esp red) from the other!
We have more recent pages around our P1 activities... but this next should still be correct.
Here is the current list of strains in the lab.
Here the current list of P1 lab users.
If you have a project idea for the lab, make sure to get trained!
Talk to the biosafety officer, Rachel Aronoff, and deputy biosafety officer, Olivier Emery, about your plans in advance!
Documentation on the wiki is highly encouraged.
Abbreviations
Biological safety level 1 lab (P1), biosafety officer (BSO), standard operating procedure (SOP)...
Microbes
We now have the right to work with organisms classified as BSL1 according to Swiss Law.
The first step is to check if the microbe you want to work is BSL1, go to https://www.bafu.admin.ch/bafu/fr/home/themes/biotechnologie/publications-etudes/publications/classification-des-organismes.html to find the official Swiss classification.
The second step is to borrow the microbe from someone that already used it before. All BSL1 manipulations are announced to the government that makes the database freely accessible here: http://www.ecogen.ch/ecogen/Forms/Register/RegisterSearch.aspx go and find a researcher. Talking with someone that has experience on that specific microbe will be useful also to get some tips and tricks on growing conditions, etc..
If nobody worked with the microbe before. It is possible to purchase a culture from the Culture Collection of Switzerland (CH): https://www.ccos.ch/
Additional repositories:
- Algae and protozoa (UK): https://www.ccap.ac.uk/
- Eukariotic cells and microbes (US):ATCC https://www.lgcstandards-atcc.org/?geo_country=ch Note: the ATCC BSL classification is American. It may differ from the Swiss one. Check the Swiss one before!
- Czech Collection of Microorganisms (CCM) http://www.sci.muni.cz/ccm/index.html
Primers
- Microsynth : ~ 10.-/30 bp primer
- Eurofins
- IDT
Sequencing
- GATC
- Eurofins Genomics
Genes
A strain carrying the full luciferase operon, and a strain carrying a GFP plasmid induced by arsenic, and a constitutive mCherry plasmid, are the first being grown in the P1 lab (1nov17).
A synthetic biology plasmid made by a golden gate multipart ligation is the next strain expected. (to make a gfp plasmid become blue...)
Following Norms
Define the planned experiments (Daniel for HTGAA Academy, Rachel bio-luminescence)
Describe training program "Intro to Biosafety"
A Hackuarium Drive folder with various handouts for the 1.5h intro course, and further biosafety information contains much of the information about the P1 course. Additionally, a prezi on basic laboratory safety is recommended viewing before the course.
P1 training includes theoretical and practical aspects of basic safety and aseptic techniques.
Lists of P1 users and engineered or synthetic strains will be kept in the same shared 'Drive' folder.
The first training is being held 6 October 2017, and notification of activities to Swiss authorities will follow as final lab details are sorted (ventilation of fume hood, water to sink).
In addition to fulfilling this training, project details must be documented in the wiki, and users must also sign a waiver and acknowledgement of risks and liability before starting any experimentation in the P1 lab.
(folder of signed waiver scans to be put in the shared folder also)
Generalized procedure
These procedure will be implemented in the lab as first benchmark in order to have a working basis.
The "project request" procedure will be a way to ensure complete transparency and applicability. It will have to be filled before any project can start by any member willing to perform GMO activities.
They are optimized for cost & material efficiency.
Maximal Culture Size
Technically the space does not allow us yet to use flasks over a volume of 500ml (incubator volume). A culture grows well at a fifth of the total volume (oxigenation requisit) which theoretically makes it 100ml culture. Safety wise, using a rather small enclosed space, spills have to stay managable. To avoid bacterial spread outside of the enclosure a maximal volume of populated medium of 200ml per flask is allowed. A maximal volume of 1L in total, populated or sterile medium, is allowed.
Spill management
Droping a vial containing liquid is common. In a lab if not prpoerly handled a spill can have more dramatic effects than droping your milk at home. Here we depict the SOP for the 4 types of spills that happen with their respective anti-spill kits; acidic, basic, contaminated and neutral spills.
- 1. Alert your collaborators
- 2. Unplug and turn-off all proximate electric equipment.
- 3. Follow the procedure describing your situaion:
Acidic spill
Mercury Spill
Project Request
Develop a document/wiki page as template where people will have to explain the GMO they want to produce.
As an idea it could be a form or a wiki page that anybody could see structured as the following
- Goal:
- - What is the purpose of the GMO / protein you want to create OR: What question should it help you answer
- ex: - Sense endocrine disruptors / Produce methane / Produce dyes
- - What is the purpose of the GMO / protein you want to create OR: What question should it help you answer
- How does it help reach your goal
- ex: - This enzyme is known to biochemically produce red dye (add source)
- ex: - The combination of protein A,B&C could produce a sensor...
- Plasmid
- - Tell which plasmid you will use (for archive purpose)
- - Give the code of the cassette you will use
- - Give the peptide translation of such cassette
- Template
- - If you want to use several building blocks for you proteins, give details on the templates
- Primers
- - Give the primers you are going to use to realise your clones
Storage and Nomenclature
A recurrent behavior, often observed in institutionalized spaces, where the followup on ones item naming and storage procedure is inexistant, is the massive loss of time to find a special item belonging to a past member. Here we describe a strict naming procedure for items that is, we think, unambiguous and still logical.
Original Timeline (circa 2016)
The original timeline had the following steps:
Mise en place équipe travail + évaluation des points critiques (cout, autorisations) 1 mois
Mise en place de laboratoire + obtention du materiel 1 mois
discussion avec les autoriteés et obtention des autorisations 2 semaines
Critical Points
What we need to sort out before we start:
This scientifically enriching, yet delicate step, is now in motion but in order to start practicing several items involving legal, technical and community topics have to be settled. Indeed, our laboratory practices and transparency as a citizen lab have to be absolutely irreproachable. The challenges we faced are described in this document.
Regarding Manuela Ocaña (Collaboratrice scientifique, PhD, Département fédéral de l'intérieur DFI, Office fédéral de la santé publique OFSP):" Les instituts, les entreprises et les organisations qui utilisent des organismes génétiquement modifiés, pathogènes ou exotiques en milieu confiné sont ainsi tenus de notifier leur activité (classes 1 et 2) ou de demander une autorisation (classes 3 et 4)." Nous allons pas depasser la classe 2, du coup nous avons besoin de faire une notification (voir dessous)
Announce the first activities
It was done with this form here
Will the enclosure be satisfactory?
We now have a perfectly closed room with 2 doors and keys.
Do we need a bio safety officer? Does this person need a specific training?
Yes, Rachel has set up a P2 lab before. She will nevertheless do another round of training and she designed an initial training program for users.
Risk assessment
- The biological risk assessment will be made through the form each member has to fill when he creates a new genetically modified strain to ensure no pathological or harmful gene is inserted in the micro-organism.
- Chemically, any inflammable liquid will be stored in the solvent cabinet. The acids and bases will be separated in two "geographically" remote cabinets. The bottles will be stored close to ground level in a resistant container able to contain the full volume of the bottles it contains to avoid spillage.
Waste management
- -> OK (see below)
Federal Coordination Centre for Biotechnology (where you have to notify level 1 activities with GMOs):
http://www.bafu.admin.ch/biotechnologie/01744/01745/index.html?lang=en
BSO-Curriculum (courses for biosafety officers):
http://www.bafu.admin.ch/biotechnologie/01744/02964/index.html?lang=en
Cantonal authority (Vaud, contact persons)
-Isabelle Dessaux (isabelle.dessaux@vd.ch )
-Olivier Gianina (olivier.gianina@vd.ch )
Our (very helpful) contact is:
Manuela Ocaña
Collaboratrice scientifique, PhD
Département fédéral de l'intérieur DFI
Office fédéral de la santé publique OFSP
Unité de direction Santé publique
Schwarzenburgstrasse 157, 3003 Berne
Tél. +41 58 462 63 66
Fax +41 58 462 62 33
manuela.ocana@bag.admin.ch
www.bag.admin.ch
Her initial email was as follow:
Bonjour,
L’office fédéral de la santé publique (OFSP) est, en collaboration avec l’office fédéral de l’environnement (OFEV), responsable pour les aspects de sécurité biologique en Suisse. L'ordonnance sur l'utilisation des organismes en milieu confiné (ordonnance sur l’utilisation confinée, OUC ; RS 814.912) règle l'utilisation d'organismes génétiquement modifiés, pathogènes ou exotiques en milieu confiné (par ex. laboratoires de recherche, de diagnostic ou d’enseignement et développement). Cette ordonnance a pour but de protéger l’être humain, les animaux et l'environnement des menaces et atteintes possibles résultant de l’utilisation en milieu confiné de tels organismes. Les instituts, les entreprises et les organisations qui utilisent des organismes génétiquement modifiés, pathogènes ou exotiques en milieu confiné sont ainsi tenus de notifier leur activité (classes 1 et 2) ou de demander une autorisation (classes 3 et 4).
Nous avons pris connaissance, avec grand intérêt, des informations concernant les activités et prestations proposées par votre laboratoire « Hackuarium », qui pourraient par ailleurs tomber dans le champ d’application de l’OUC. En effet, et selon le type de matériel utilisé (par ex. organismes génétiquement modifiés ou pathogènes) ainsi que la nature et l’ampleur de vos activités, ces dernières pourraient être soumises au devoir de notifier selon l’OUC. Mais avant d’entreprendre des démarches qui pourraient s’avérer finalement inutiles, je vous propose de consulter d’abord les informations disponibles sur le site du Bureau de Biotechnologie de la Confédération puis de reprendre contact avec nos services afin de clarifier au mieux votre situation.
N’hésitez à me contacter en tout temps si vous avez des questions ou besoin d’informations supplémentaires.
Pivot Points
Here we develop and tackle down the most critical points of this project. The idea is to estimate if our space can accommodate such an infrastructure and if our community has the shoulders to carry the necessary responsibilities. The points have to be assessed keeping in mind the first standardized lab procedure that will be globally used by the lab (see generalized procedure).
Lab room and material
Here we discuss the size features and needed characteristics of the room we are going to use for the lab. We also describe a material list we would need to equip the lab to the minimum.
Room features
Material
All the details for the material we would need is on the following doc: https://docs.google.com/spreadsheets/d/1tBvxEk4UVtvzfeW_-yR9yNbrLmC1V9IDjAPit37TzAg/edit#gid=1606970814
Material to be used for HTGAA 2017 is here: https://docs.google.com/spreadsheets/d/1Xg_GtrFb8wYJVUPlrbUb6pP0dWGmkoAKU4xP6qhBKVE/edit#gid=0
Waste elimination
Here we describe who can and accepts to eliminate our wastes at what cost and try to estimate the frequency of elimination. We base the frequency on a per project base. How many liters of cultures are produced per production? How many biowaste bag can be filled in a week? etc...
Procedure
Based on: http://www.cusstr.ch/repository/138.pdf
- Les souches non pathogènes et non modifiées génétiquement ne nécessitent pas d’inactivation. Les déchets liquides peuvent être éliminés dans l’évier pour autant qu’il n’y ait pas d’autres contaminants p.ex. chimiques ou radioactifs. Les déchets solides doivent être éliminés avec les déchets incinérables, avec les mêmes restrictions que les liquides.
- Tout matériel de laboratoire contaminé par un OGM de classe 1 doit être inactivé avant élimination pour une filière de déchet normal. Un marquage clair des sacs à déchets biologiques doit être effectué afin de pouvoir identifier aisément les déchets autoclavés de ceux qui ne l’ont pas encore été. Par exemple, le sac auroclavé sera transféré dans un autre sac de couleur différente ne laissant plus apparaître le sigle biohazard et le contenu.
- Les objets tranchants ou coupants doivent être éliminés comme déchets spéciaux.
In english in a sentence Bio-waste bags have to be clearly distinguishable from other waste bags for example with the bio-hazard sign on them. Once inactivated (i.e. autoclaved) you have to package the waste in a way that it neither doesn't look nor doesn't smell disgusting or dangerous. Then you can discard it as household waste. If this is not possible, it needs to be discarded as special waste, how it is done at EPFL (white bag with red stripes).
Service providers
Since the bags of solid wastes can be eliminated through the classical way and the autoclaved liquids can be poured to the toilets the cost can be estimated negligible.
No need for special waste removal
Calculated base costs
- Here we estimated the cost of starting the whole setup, based on the output of a reference protein laboratory's costs.
- We also took a standardized method which is documented lower (protocol) based only on biology with E.coli.
- The costs are detailed on the doc below and calculated on the academic standards (ca. 2015 calculations).
- The goal then is to find hacks and shortcuts to largely cut the costs and make it affordable.
- This exercise still needs to be redone for the 2017 P1 context.
https://docs.google.com/spreadsheets/d/1tBvxEk4UVtvzfeW_-yR9yNbrLmC1V9IDjAPit37TzAg/edit#gid=0
Software and Databases
Here we discuss the gene editing softwares to design plasmids, primers and gene building blocks. A total transparency has to be implemented in order for people to follow our activities. What kind of database will we use to store the ordered primers sequences? Same for the plasmids?
Software
- Geneious powerful but the file format is not opensource or genebank
- Snapgene never tried, anyone ?
- One of the more recent (oct2017) on-line tools for molbio work is called Benchling, which can also be used for an on-line lab notebook. This tool is currently being used in the context of the HTGAA course.
Database
Providers
Here we discuss the possible service providers, their costs and if they are ready to deal with us. We canot create plasmids without primers or genes and therefore need providers. We cannot confirm the authenticity of our work without a verified plasmid sequence and therefore need sequencing services.
Legal
Backup of the first exploration document : http://wiki.hackuarium.ch/w/Talk:Pushing_the_P1#Old_Hackpad_Page (Most of the necessary infos can be found here)
As said previously our laboratory practices have to be irreproachable. We carry the "hacker" label and therefore, for the public opinion and the built of trust. We need to study in depth several aspects of the legislation governing genetic manipulation.
WHO
The very good lab safety manual right here
Go through it if you have the time or are curious, safety in the lab is everybody's job.
Swiss legislation
On Feb 3 2016, we got some pointers from the Vice Dean of the School of Biology of UniL who passed us information from Audvion.ch (Ingénieur de sécurité MSST)
From: Objet: AVP réponse biosécurité Re: Laboratoires P1 Date: 3 février 2016 13:58:56 UTC+1
Pour toutes les questions de sécurité biologique (biosécurité) les règles de base sont données dans deux ordonnances doit :
- OUC Ordonnance sur l'utilisation des organismes en milieu confiné https://www.admin.ch/opc/fr/classified-compilation/20100803/index.html
- OPTM Ordonnance sur la protection des travailleurs contre les risques liés aux microorganismes https://www.admin.ch/opc/fr/classified-compilation/19994946/index.html
Vous pouvez aussi consulter
- Le site de la CUSSTR (Commission universitaire pour la santé et la sécurité au travail romande) http://cusstr.ch/fr/doc/technique/detail/?idcat=14
- Un petit aide mémoire de la SUVA sur le sujet (missing link)
- Vous référer au coordinateur de biosécurité BSO de votre département
On Feb 25 2016, we got some more pointers from the Office fédéral de la santé publique OFSP
L’office fédéral de la santé publique (OFSP) est, en collaboration avec l’office fédéral de l’environnement (OFEV), responsable pour les aspects de sécurité biologique en Suisse. L'ordonnance sur l'utilisation des organismes en milieu confiné (ordonnance sur l’utilisation confinée, OUC ; RS 814.912) règle l'utilisation d'organismes génétiquement modifiés, pathogènes ou exotiques en milieu confiné (par ex. laboratoires de recherche, de diagnostic ou d’enseignement et développement). Cette ordonnance a pour but de protéger l’être humain, les animaux et l'environnement des menaces et atteintes possibles résultant de l’utilisation en milieu confiné de tels organismes. Les instituts, les entreprises et les organisations qui utilisent des organismes génétiquement modifiés, pathogènes ou exotiques en milieu confiné sont ainsi tenus de notifier leur activité (classes 1 et 2) ou de demander une autorisation (classes 3 et 4).
Nous avons pris connaissance, avec grand intérêt, des informations concernant les activités et prestations proposées par votre laboratoire « Hackuarium », qui pourraient par ailleurs tomber dans le champ d’application de l’OUC. En effet, et selon le type de matériel utilisé (par ex. organismes génétiquement modifiés ou pathogènes) ainsi que la nature et l’ampleur de vos activités, ces dernières pourraient être soumises au devoir de notifier selon l’OUC. Mais avant d’entreprendre des démarches qui pourraient s’avérer finalement inutiles, je vous propose de consulter d’abord les informations disponibles sur le site du Bureau de Biotechnologie de la Confédération (http://www.bafu.admin.ch/biotechnologie/01744/01745/index.html?lang=fr ) puis de reprendre contact avec nos services afin de clarifier au mieux votre situation.
BioSafety Officier (BSO)
a few members from the community did the course below...
Courses : http://www.bafu.admin.ch/biotechnologie/01744/02964/index.html?lang=f
Infos:
BSO, niveau de sécurité 1 (BSL1_2016)
Date: 9 septembre 2016 (1 jour)
Lieu: Université de Berne
Inscription d'ici au 30 juin 2016 à: registration@curriculum-biosafety.ch
Coûts: 550 francs par personne
Langue: anglais
update 2017, RA will be BSO for new P1, train new users, and sign up for a refresher course soon.
She has made some edits on the P1 lab SOP aka Standard Operating Procedure (still a work in progress) and did the notification of activities!
Annonce confédération
Through the BAFU portail
Train the users
Put it a plan that authorizes the new people to come in
Write the notification to the Biosafety Authorities
Users
Oppositely to the Hackuarium's "P0" lab where users can come and experiment in the greatest of liberties the P1, as previously stated, will be restricted to card holders having followed an initiatory course. Each user will be registered in a database and will chose a three letter OR digit code to identify items made by himself. Example:
- John Doe : joh
- Martin Luther King : mlk
- George Junior : gj2
This three letter acronym is practical as it is short enough to be stuck on small tubes with a descriptor i.e: joh_p7 (John Doe Plasmid 7)
Descriptors
In the process of producing for example a protein from scratch (from synthetic DNA) only a few items have to be stored and conserved for further use. These items often in the forms of solutions rarely exceed volumes of 200ul. Therefore the whole process of protein engineering and plasmid creation can hold in 1.5ml tubes stored in 96well cardboard freezer boxes. We list what we extimate should be conserved for further uses by other collaborators and give them a single letter code.
- project/task/job : j
- DNA segments : d
- primers/amorces : a
- plasmids : p
- proteins/constructs : c
- bacteria/strain : b
These six items are the valuable items to be stored. The project (j) item might seem counter intuitive but makes sense in the whole nomenclature system. Let's give some examples with the hypothetical John Doe project 41 (i.e joh_j41).
- He might do a plasmid (joh_j41_p1) that sadly he failed so he produced four more for sequencing (joh_j41_p2, joh_j41_p3, joh_j41_p4, joh_j41_p5).
- The plasmid was a construction of a vector (joh_j41_dV) and two inserts (joh_j41_dI1, joh_j41_dI2)
- The latter were made with six primers (joh_j41_a1, joh_j41_a2, joh_j41_a3, joh_j41_a4, joh_j41_a5, joh_j41_a6)
- He finally made it and expressed his protein (joh_j41_c1) (Usually you only make a protein from a plasmid so if you re-express, as a 3rd batch, it later on: joh_j41_c3)
The nomenclature might appear cumbersome at some point but it should make more sense once the final project draft is presented.
Storage
When you're in the working process, the following behavior is observed: You open the freezer, look for the box containing the items of interest (generally you have a primer box, a plasmid box, etc.), open the box and in those 96 tubes you want to find them by looking at the top of them. The "morphology" of a 1.5ml tube gives you only two options: You can write on the top only limited information. Forget about markers they will fade away unless you add transparent tape on top (which is boring). Or you can add stickers on the side of the tube that can contain more details on the content. So the following storage procedure will be observed:
- Boxes will only contain a single type of items such as primers, plasmids or proteins.
- Each item of whatever sort will have a unique 4 letter/digit hex code preceded by the type of item. That's roughly 65500 possibilities. This code is preceded by the adequate descriptor. Example: joh_j41_p1 (the plasmid N°1 of John's project 41) was given by the system the code p019A this is 5 letters which perfectly fit on a 1.5ml tube top if printed on a sticker. Again this looks cumbersome know but will make more sense later since the system gives and stores all the info for you.
Job page
We presented previously the request form. This form generates for you a wiki project page or whatever online lab-book that we still have to find. The final idea is that on the server the access to these code and data is easily accessible from a terminal in the P1 lab.
Here is an hypothetical project job page and how it should look like in the end after the form has been filled.
When somebody works and experimentally gets the product design then he can directly fill the page or the online lab-book that should transfer the data.
A final page, with experimental data filled in should look like this
If a page is filled with such details, reproducibility and experimental follow-up is much easier. The important point is having a strong automation of the main points so as to avoid time loss of creating manually those files (took me more than 30 min making a complete one by hand, it should not take more than 10 and should be done while doing the experiment).
» Project request form with above points is now here.
Bacterial
All of these components have to be open source.
Strain: e.coli
Plasmid:
Resistance: Ampicilin
DNA Amplification / Cloning
- Good old openTaq
- DNA fragment purification by gel extraction. (cheap, easy, no need of DPNI enzymes)
- Ligation through modified Gibson (lysed bacterial supernatant + enzymatic complementation (Exonuclease mostly), To investigate further see the SLiCE method)
- Transformation in chemo competent cells by heat shock (way cheaper than electro-competent cells)
- Usual LB-agar plating with ampicilin selection
- How do we purify the DNA from the cultures ? (yeah of course we can use qiagen minipreps but it's expensive...) There is an easy spermidine DNA prep that works all the times and costs nothing.
Primers production
Can we inspire ourselves from the cheap DIY peptide synthetiser and turn it to a primer systhetiser ? Source: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4045704/#!po=52.8409
Cultures
- e.coli chemo competent cells will be stored in a -80°C fridge.
- Transformation will be made by heat shock as described in ("doc name here").
- Classical LB-Amp plating with static overnight 37°C incubation.
- Liquid cultures can be either made as:
- LB-Amp followed by IPTG induction.
- (-) IPTG is expensive
- (-) Monitoring of optical density requires time + UV-spectro
- (-) Large volumes (1L medium in 5L flasks) for decent yields + large capacity incubator cooling to 16°C = more waste
- (-) Have to centrifuge in 500ml flasks = more post-culture work.
- (+) yields are often good, procedure is bulletproof
- Modified auto-inducible Terrific-Broth (TB-of-doom)
- (-) more complex medium preparation (can still be made as large stock)
- (+/-) yields are good but proteins might be hard to extract
- (+) No UV monitoring ("by eye check"), after growth expression can be made at room temp.
- (+) small 100ml cultures in 500ml flasks
- (+) all post-culture fork can be made in 2x50 ml flasks (convenient)
Protein purification
- Purification can be made by single His tag purification
- We have more than 200ml of Ni-NTA resin
- The resin can be recycled easily
- Quite simple procedure (requires 3 buffers)
- (-) Single His-tag purif will not yield absolutely pure proteins
- Concentration
- problem remains to be solved
- Storage will be done at -20°C in 40% glycerol
Yeast
Yeast specialists please provide a protocol applicable here, keep in mind cost & material optimization.
Old Plan for P1 from 2016 - OP Greenhouse Effect
Building the P1 in a green house... I can already smell ideas fresh as spring flourishing in there...
Idea
This clever and original idea was brought to us by our master of "let's just do it it's gonna work anyways" Gustavo, congratulation to him!
Therefore we can buy a greenhouse for a reasonable price and set it up as a P1. The size will be indeed smaller but will be enough to accomodate the transformation, growth, cell lysis and waste storage until elimination.
Several greenhouses models are presented with a rationale on which one to pick.
The end of the document is supplemented with tasks and points to investigate
Greenhouses
We indeed aim for the best size/price ratio.
- Serre Delia, Hornbach, 11m2, 1399.- link
- Initial idea
- (-) Expensive
- (+) Higher at the wall/roof junction (1m40) than the others. Still 1m40 is not very high.
- Serre Vida XL, 12.25m2, VidaXL, 399.- link
- (+)Free delivery
- (+)Cheap
- (-)Low at the wall/roof junction (95cm). BUT! For the price we can get 2 and add the two walls on top of the other giving 1.9m!.
==Building Plan==
(the updated Draft is above, a complete version of this with measure a nice pinky shinny colors will follow the next days)
Tasks
General
- 1. Members validate the idea or propose other ideas until Wed. (DONE)
- 2. Ordering and building all together on the 2nd of Oct. (MATERIAL RECEIVED)
- 3. Building zone cleaned (DONE)
Material & Orders
- 1. Rearrange lab space (DONE)
- 2. Think about a list of vital chems and consumables that are needed and could be stored in the P1 for easier access.
- look at: https://docs.google.com/spreadsheets/d/1tBvxEk4UVtvzfeW_-yR9yNbrLmC1V9IDjAPit37TzAg/edit#gid=0 and select the vital items.
- 3. Get labcoats of various sizes, get hooks for those
- 4. Get a set of safety goggles
- 5. Find fridge + -20 freezer
- 6. Order desinfectant dispenser
- 7. Get 3x6 multiplugs
- 8. Card access system ( can use yann's raspi3)
- 9. Get sturdy, closable bin for solid biowaste
- 10. Bin for classic wastes
- 11. Sam, get a hell stock of printable stickers (we're gonna label the shit out of everything)
- 12. Organize one cupboard with:
- pcr purif kits
- miniprep kits
- gel purif kits
- 50, 12 & 1.5 ml plastic tubes
- 1, 200 & 1000ul tips
- his tag purification buffers
- 13. On the sterile hood, find a plexiglass that we can fix on and that will cover 2/3 of the front pannel (i.e. Will make a separation betwwen the user and the working environement
- 13. Order a pipetor on SMIPLES / ricardo for serological pipettes
- 14. We have two benches with the glass tops: one is not held with screws, fix it, buy long enough screws (obi)
- 15. Clean a set of micropipettes
Safety
- 1. Announce our first activity on the confederation website (Yann)
- 2. Design the safety standard operating procedures (SOP) such as:
- MSDS for common chemicals
- What to do if a spillage of contaminated liquid occurs (how to clean it).
- What to do if you get injured working with bacteria.
- How to eliminate wastes, liquid and solid.
- Standard lab behavior (blouse, glasses, gloves)
- Where to store: acids, bases, solvents -> Rules for using each of them (how bases and acids have to be separated separately)
- A very good example : http://ehs.berkeley.edu/standard-operating-procedures
- 3. Print the following decals
- Biohazard 2x
- Authorized personnel only
- "Wash your hands when done"
- "Max X users inside" (X to be defined)
- 4. Find an easy an accessible way to put blouses and glasses (gloves can be stored and boxes left above the bench)
- 5. Make instrument procedures (print and put in a folder)
- 6. Questions? There is a service in the DIYBio community for Ask a Biosafety Professional
Ideas & Concerns
- As the name says it's a greenhouse, how do we make sure it doesn't become super warm ? Can we make a cheap air cycling system ?
Lab Setup
An older idea: Current setup higher up on this page