Pushing the P1
Introduction
Having a lab space and instruments is one thing, possessing the tools to do real biology is another thing. For close to two years now, the Hackuarium community has built a space, where people can work on projects related to biology among other fields. The pretension of the international DIY bio scene, or closely related biohacking scene, is to bring to the people the tools to perform and work on the whole range of biological applications, as qualitatively and cheaper than what the industrial or academic institutions do. To achieve such level, the Hackuarium lab has to be upgraded to a higher level of competency. This level is the P1 biosafety level where genetic manipulation open the door to a vast range of research topic and engineering opportunities. This scientifically enriching, yet delicate step, is now in motion but in order to start practicing several items involving legal, technical and community topics have to be settled. Indeed, our laboratory practices and transparency as a citizen lab have to be absolutely irreproachable. The challenges we face are described in this document.
We will prepare for the P1 work assuming it will start out in bacteria, and then depending on future demands, we imagine adding other organisms.
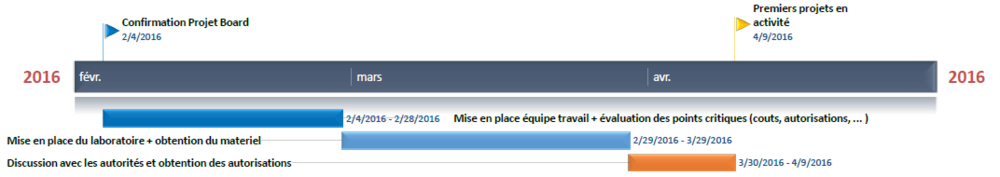
Timeline
Here is the basic timeline we will try to keep in order to make the project move on. It is obviously a "best case scenario" approximation.
Critical Points
- will the enclosure be satisfactory? (legally)
- do we need a bio safety officer ?
Pivot Points
Here we develop and tackle down the most critical points of this project. The idea is to estimate if our space can accommodate such an infrastructure and if our community has the shoulders to carry the necessary responsibilities. The points have to be assessed keeping in mind the first standardized lab procedure that will be globally used by the lab (see generalized procedure).
Lab room and material
Here we discuss the size features and needed characteristics of the room we are going to use for the lab. We also describe a material list we would need to equip the lab to the minimum.
Room features
Material
Stockage:
- Congelateur -80°C, -20°C: -50, 60C freezer used for tuna transport comes in a smaller size.
- Frigo, 4°C
- Glace
Transformation:
- Heatblock statique
- Heatblock shaker
Culture:
- Incubateur statique
- Incubateur rotatif (pouvant refroidir)
Post-culture:
- Centrifuge large volume
- Ultra centrifuge
- Sonication
- Centrifuge 1.5ml
- Autoclave destruction
Mobilier:
- Benches
- Armoires
Autres:
- Bec Bunzen
- Sturdy trash box
- Safe liquid waste flask
Waste eliminations
Here we describe who can and accepts to eliminate our wastes at what cost and try to estimate the frequency of elimination. We base the frequency on a per project base. How many liters of cultures are produced per production? How many biowaste bag can be filled in a week? etc...
Costs
Service providers
Frequency
Calculated base costs
Here we estimate the cost of starting the whole setup. Taking a standardized transfection method, expression method, cell line and antibiotic for the whole lab, how much would it cost to start it up? What are the most important enzymes? Are there opensource alternatives?
Enzymes / Plasmids / Competent cells
Cultures
Wastes
Chemicals
Summary
Softwares and Databases
Here we discuss the gene editing softwares to design plasmids, primers and gene building blocks. A total transparency has to be implemented in order for people to follow our activities. What kind of database will we use to store the ordered primers sequences? Same for the plasmids?
Softwares
Database
Providers
Here we discuss the possible service providers, their costs and if they are ready to deal with us. We canot create plasmids without primers or genes and therefore need providers. We cannot confirm the authenticity of our work without a verified plasmid sequence and therefore need sequencing services.
Primers
Sequencing
Genes
Legal
Hackpad first exploration document : https://hackpad.com/Wanna-go-P1--I8F1GOCYTR9 (Most of the necessary infos can be found here)
As said previously our laboratory practices have to be irreproachable. We carry the "hacker" label and therefore, for the public opinion and the built of trust, cannot fuck around. We need to study in depth several aspects of the legislation governing genetic manipulation.
WHO
Swiss legislation
On Feb 3 2016, we got some pointers from the Vice Dean of the School of Biology of UniL who passed us information from Audvion.ch (Ingénieur de sécurité MSST)
From: Objet: AVP réponse biosécurité Re: Laboratoires P1 Date: 3 février 2016 13:58:56 UTC+1
Pour toutes les questions de sécurité biologique (biosécurité) les règles de base sont données dans deux ordonnances doit :
- OUC Ordonnance sur l'utilisation des organismes en milieu confiné https://www.admin.ch/opc/fr/classified-compilation/20100803/index.html
- OPTM Ordonnance sur la protection des travailleurs contre les risques liés aux microorganismes https://www.admin.ch/opc/fr/classified-compilation/19994946/index.html
Vous pouvez aussi consulter
- Le site de la CUSSTR (Commission universitaire pour la santé et la sécurité au travail romande) http://cusstr.ch/fr/doc/technique/detail/?idcat=14
- Un petit aide mémoire de la SUVA sur le sujet (missing link)
- Vous référer au coordinateur de biosécurité BSO de votre département
BioSafety Officier (BSO)
Annonce confédération
Norms
Generalized procedure
These procedure will be implemented in the lab as first benchmark in order to have a work basis.
The "project request" procedure will be a way to ensure complete transparency and applicability. It will have to be filled before any project can start by any member willing to perform GMO activities.
They are optimized for cost & material efficiency.
Project Request
Develop a document/wiki page as template where people will have to explain the GMO they want to produce.
As an idea it could be a form or a wiki page that anybody could see structured as the following
- Goal:
- - What is the purpose of the GMO / protein you want to create OR: What question should it help you answer
- ex: - Sense endocrine disruptors / Produce methane / Produce dyes
- - What is the purpose of the GMO / protein you want to create OR: What question should it help you answer
- How does it help reach your goal
- ex: - This enzyme is known to biochemically produce red dye (add source)
- ex: - The combination of protein A,B&C could produce a sensor...
- Plasmid
- - Tell which plasmid you will use (for archive purpose)
- - Give the code of the cassette you will use
- - Give the peptide translation of such cassette
- Template
- - If you want to use several building blocks for you proteins, give details on the templates
- Primers
- - Give the primers you are going to use to realise your clones
» Project request form with above points is now here.
Bacterial
All of these components have to be open source.
Strain: e.coli
Plasmid:
Resistance: Ampicilin
DNA Amplification / Cloning
- Good old openTaq
- DNA fragment purification by gel extraction. (cheap, easy, no need of DPNI enzymes)
- Ligation through modified Gibson (lysed bacterial supernatant + enzymatic complementation (Exonuclease mostly), To investigate further)
- Transformation in chemo competent cells by heat shock (way cheaper than electro-competent cells)
- Usual LB-agar plating with ampicilin selection
- How do we purify the DNA from the cultures ? (yeah of course we can use qiagen minipreps but it's expensive...) There is an easy spermidine DNA prep that works all the times and costs nothing.
Primers production
Can we inspire ourselves from the cheap DIY peptide synthetiser and turn it to a primer systhetiser ? Source: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC4045704/#!po=52.8409
Cultures
- e.coli chemo competent cells will be stored in a -80°C fridge.
- Transformation will be made by heat shock as described in ("doc name here").
- Classical LB-Amp plating with static overnight 37°C incubation.
- Liquid cultures can be either made as:
- LB-Amp followed by IPTG induction.
- (-) IPTG is expensive
- (-) Monitoring of optical density requires time + UV-spectro
- (-) Large volumes (1L medium in 5L flasks) for decent yields + large capacity incubator cooling to 16°C = more waste
- (-) Have to centrifuge in 500ml flasks = more post-culture work.
- (+) yields are often good, procedure is bulletproof
- Modified auto-inducible Terrific-Broth (TB-of-doom)
- (-) more complex medium preparation (can still be made as large stock)
- (+/-) yields are good but proteins might be hard to extract
- (+) No UV monitoring ("by eye check"), after growth expression can be made at room temp.
- (+) small 100ml cultures in 500ml flasks
- (+) all post-culture fork can be made in 2x50 ml flasks (convenient)
Protein purification
- Purification can be made by single His tag purification
- We have more than 200ml of Ni-NTA resin
- The resin can be recycled easily
- Quite simple procedure (requires 3 buffers)
- (-) Single His-tag purif will not yield absolutely pure proteins
- Concentration
- problem remains to be solved
- Storage will be done at -20°C in 40% glycerol
Yeast
Yeast specialists please provide a protocol applicable here, keep in mind cost & material optimization.
Teams & Tasks
- Costs
- Providers
- Softwares
- Waste handling
- Legal
- Yeast protocol
- Form