Difference between revisions of "Openhackuarium vol. 115"
(→Links) |
|||
| (2 intermediate revisions by the same user not shown) | |||
| Line 6: | Line 6: | ||
| − | == IntelliGene | + | == iGEM project: IntelliGene == |
| + | |||
| + | iGEM (international Genetically Engineered Machine) is an organization and communty “dedicated to education and competition, advancement of synthetic biology, and the development of open community and collaboration”. Every year EPFL hosts a team which goes to Boston to compete in the Giant Jamboree, a massive international competition for synthetic biology projects. Projects are displayed in poster, presentation, and web-based formats, and submitted to panel of judges who evaluate them for medal requirements such as the creation of novel biological constructs, the improvement of previous biological constructs, and community engagement. '''This years team, IntelliGene, will give us a sneak preview of their project before going to Boston!''' | ||
| + | |||
| + | === Project Description === | ||
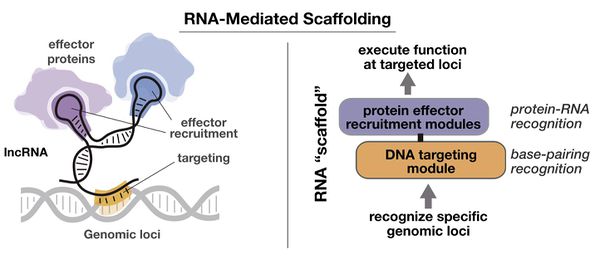
Designer cells are becoming ever more important in the world of synthetic biology. To be able to conveniently harness this technology, new and straightforward tools are required. In light of this, our project aims to develop an innovative CRISPR-dCas9 system in yeasts, capable of regulating genetic transcription and creating robust synthetic circuits. Our model is based around a scaffold guide RNA. This scaffold allows us to recruit transcriptional activators, repressors, and dCas9, as well as direct the complex to a given locus in the genome. In addition, the presence of both activators and repressors in our system would permit a modularity previously unseen in CRISPR-dCas9 based systems. Furthermore, we aspire to improve on Cello, a software that takes a user-given circuit and predicts a plasmid that could recreate it in vivo. | Designer cells are becoming ever more important in the world of synthetic biology. To be able to conveniently harness this technology, new and straightforward tools are required. In light of this, our project aims to develop an innovative CRISPR-dCas9 system in yeasts, capable of regulating genetic transcription and creating robust synthetic circuits. Our model is based around a scaffold guide RNA. This scaffold allows us to recruit transcriptional activators, repressors, and dCas9, as well as direct the complex to a given locus in the genome. In addition, the presence of both activators and repressors in our system would permit a modularity previously unseen in CRISPR-dCas9 based systems. Furthermore, we aspire to improve on Cello, a software that takes a user-given circuit and predicts a plasmid that could recreate it in vivo. | ||
Latest revision as of 14:06, 26 August 2016
Main language this week: English
Theme : Building tools for easy synthetic genetic circuit creation
Date : Wednesday, September 14th, 2016. Doors open at 7.30pm, presentations at 8 pm
iGEM project: IntelliGene
iGEM (international Genetically Engineered Machine) is an organization and communty “dedicated to education and competition, advancement of synthetic biology, and the development of open community and collaboration”. Every year EPFL hosts a team which goes to Boston to compete in the Giant Jamboree, a massive international competition for synthetic biology projects. Projects are displayed in poster, presentation, and web-based formats, and submitted to panel of judges who evaluate them for medal requirements such as the creation of novel biological constructs, the improvement of previous biological constructs, and community engagement. This years team, IntelliGene, will give us a sneak preview of their project before going to Boston!
Project Description
Designer cells are becoming ever more important in the world of synthetic biology. To be able to conveniently harness this technology, new and straightforward tools are required. In light of this, our project aims to develop an innovative CRISPR-dCas9 system in yeasts, capable of regulating genetic transcription and creating robust synthetic circuits. Our model is based around a scaffold guide RNA. This scaffold allows us to recruit transcriptional activators, repressors, and dCas9, as well as direct the complex to a given locus in the genome. In addition, the presence of both activators and repressors in our system would permit a modularity previously unseen in CRISPR-dCas9 based systems. Furthermore, we aspire to improve on Cello, a software that takes a user-given circuit and predicts a plasmid that could recreate it in vivo.
Links
https://www.facebook.com/igemepfl
https://twitter.com/EPFL_iGEM
Practical Info (access, map, etc.)
cf. #OpenHackuarium